 It’s nearing the end of #DepositorWeek at Addgene and we’d like to show our gratitude on the blog by highlighting some of the great posts written by Addgene depositors. Below you’ll find brief summaries of several excellent guest posts from our depositors. Interested in writing your own post for the Addgene Blog? Fill out our guest blogger form to get started!
It’s nearing the end of #DepositorWeek at Addgene and we’d like to show our gratitude on the blog by highlighting some of the great posts written by Addgene depositors. Below you’ll find brief summaries of several excellent guest posts from our depositors. Interested in writing your own post for the Addgene Blog? Fill out our guest blogger form to get started!
Thanks for all your hard work!
Fluorophores
GFP oligomerization can lead to misleading artefacts in your experiments. Read this post from Erik Snapp, Associate Professor at Albert Einstein College of Medicine, to learn how problematic oligomerization can be and how to avoid it in your experiments.
 Better Dyeing Through Chemistry & Small Molecule Fluorophores
Better Dyeing Through Chemistry & Small Molecule Fluorophores
In this post, Luke Lavis from Janelia Research Campus reminds us that, although fluorescent proteins are incredibly useful, chemical fluorophores can still be used in a wide variety of experiments. Read more to find chemical fluorophores for your next experiment.
Nucleic acid based machines
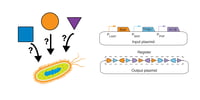 Recombinase-based State Machines Enable Order-dependent Logic in vivo
Recombinase-based State Machines Enable Order-dependent Logic in vivo
Nathaniel Roquet from the Lu Lab at MIT developed plasmid based systems that report the order in which specific chemical inputs are received by a cell. These so-called recombinase-based state machines could potentially be used to report upon and/or engineer “temporally ordered biochemical events” such as those that occur during development. Read this post to learn more about recombinase-based state machines and how they function.
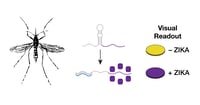 Adapting Toehold Switches to Detect Zika Virus
Adapting Toehold Switches to Detect Zika Virus
The RNA-based toehold switches described in this post are engineered to bind RNA sequences derived from Zika virus and, upon binding, produce reporter proteins. These toehold switches can be used to quickly detect the presence of Zika virus. Read this post from Keith Pardee and Alexander Green (both formerly at the Wyss Institute) to learn how these amazing tools may be used to rapidly diagnose Zika virus infection.
Microbiology and virology
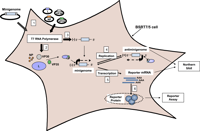 Minigenomes - A Safe Way to Study Dangerous Viruses
Minigenomes - A Safe Way to Study Dangerous Viruses
Contributed by guest blogger Tessa Cressy from the labs of Dr. Elke Mühlberger and Dr. Rachel Fearns at Boston University School of Medicine, this post describes a tool you can use to study basic virus biology while leaving virulent components behind. Minigenomes are stripped down viral genomes containing only the sequences required for viral replication and transcription as well as reporter genes. To learn how you can use minigenomes in your research, check out this post!
 R Bodies: Membrane-Rupturing Microscopic Tools
R Bodies: Membrane-Rupturing Microscopic Tools
In this post, Jessica Polka, formerly a postdoctoral researcher in the Silver Lab at Harvard University, describes R-bodies, “force generating protein machines that can extend and retract over many microns in response to pH changes without the need for ATP or other chemical fuel sources.” In her work, Dr. Polka made R-body mutants that respond to different pH’s and some that are fused to fluorescent proteins. She also showed that R-bodies can rupture E. coli membranes. Read the full post for potential applications of R-bodies and to find a video of them in action.
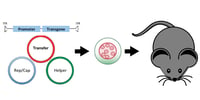 AAV: A Versatile Viral Tool for Gene Expression in Mammals
AAV: A Versatile Viral Tool for Gene Expression in Mammals
Adeno-associated viruses (AAVs) are commonly used to deliver a genes of interest to a variety of cell types in-vivo. These are widely used by the optogenetics and chemogenetics communities to activate neurons and neuronal pathways using light and chemicals respectively. Didem Goz Aytruk from Connie Cepko’s Lab at Harvard Medical School describes how AAVs function and indicates important things to consider if you’re getting started with them.
CRISPR
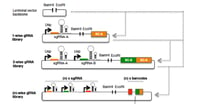 Pairing CombiGEM and CRISPR for Combinatorial Genetic Screening
Pairing CombiGEM and CRISPR for Combinatorial Genetic Screening
CombiGEM is a technology that can be used to quickly barcode a set of DNA sequences. In this post, Alan Wong (formerly of the Lu Lab at MIT) shows how you can use CombiGEM to barcode small libraries of CRISPR gRNAs to use for screening experiments. Get the full details on how to make these libraries from the post and the associated publication.
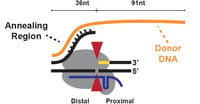 Optimizing Donor DNA for Enhanced CRISPR Genome Editing
Optimizing Donor DNA for Enhanced CRISPR Genome Editing
CRISPR has revolutionized how researchers make knockouts and other types of genome edits, but, even with CRISPR, it isn’t always easy to insert a section of DNA into the genome of your target organism. In this post, Chris Richardson, a postdoctoral researcher in Jacob Corn’s lab, describes how you can better design your donor DNA template to achieve more efficient genome editing - all influenced by an understanding of Cas9 biophysics!
 Casilio: An Adaptive, Multitasking “CRISPR-OS”
Casilio: An Adaptive, Multitasking “CRISPR-OS”
In this post, Albert Cheng and Mark Wanner from the Jackson Laboratory for Genomic Medicine described Casilio, a system that uses modular RNA binding proteins, dCas9, and a modified gRNA to direct a variety of customizable functions to your DNA sequence of interest. Potential applications of the system include activation and repression of your target gene, activation and repression of different genes in a single cell, and localization of a fluorescent signal to your target gene. Read this post to find out more about how the Casilio system works and how you can customize it for your experimental needs. Note: Casilio is no longer available through Addgene.
Thanks again to all of our fantastic depositors and guest writers. Your contributions make Addgene and the Addgene Blog useful resources for the biological research community!

Topics: Addgene News
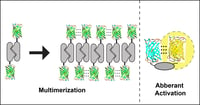 When is a Monomer not a Monomer? The Top Three Ways Your Favorite Fluorescent Protein Oligomerizes in Cells
When is a Monomer not a Monomer? The Top Three Ways Your Favorite Fluorescent Protein Oligomerizes in Cells





Leave a Comment