This post was contributed by guest blogger, Luke Lavis, a Group Leader at the Janelia Research Campus, Howard Hughes Medical Institute.
Chemistry is dead, long live chemistry!
The discovery of green fluorescent protein (GFP) sparked a renaissance in biological imaging. Suddenly, cell biologists were no longer beholden to chemists and (expensive) synthetic fluorophores. Add a dash of DNA with an electrical jolt and cells become perfectly capable of synthesizing fluorophore fusions on their own. Subsequent advances in fluorescent proteins have replicated many of the properties once exclusive to small-molecules: red-shifted spectra, ion sensitivity, photoactivation, etc. These impressive advances lead to an obvious question: In this age of GFP and its ilk, why should cell biologists talk to chemists?
One reason is something called “the photon budget”: each sample has a limited number of fluorophores and each fluorophore can emit a limited number of photons before bleaching. The amount of information one can extract from a biological sample is wholly dependent on the photon budget and pushing the frontiers of fluorescence microscopy often requires more photons. For example, moving from transient overexpression of protein fusions to gene-edited cells can decrease the number of fluorophores, compromising the photon budget. Likewise, the switch from ensemble imaging to single-molecule imaging places a greater burden on the photon budget, which determines how long and precise we can track individual molecules. Poor photon budgets are a widespread issue with fluorescent proteins—even the thriftiest cell biologist can feel like a college student searching couch cushions for spare change, desperate to extract a few more photons from a sample.
Chemical fluorophores can be substantially brighter and more photostable than fluorescent proteins, providing a straightforward way to improve the photon budget. Of course, “reverting” to small molecule dyes seems daunting—no one wants to spend their days microinjecting fluorescent conjugates into cells. Fortunately, over the last 20 years clever chemists and biochemists have developed techniques to make labeling chemistry easier and more functional in complex biological environments such as live cells and tissues (Figure 1). These flexible strategies give you the best of both worlds: the excellent photophysics of chemical dyes combined with the ease and specificity of fluorescent proteins.
In-cell labeling strategies
The majority of in-cell labeling strategies have two parts: (1) a genetically encoded “tag” expressed as a fusion with your favorite protein and (2) a synthetic fluorophore-contianing “ligand” that binds to the tag. Like most good ideas in biological imaging, the initial breakthrough for in-cell labeling techniques was provided by Roger Tsien, who showed that the selective interaction between a bisarsenical dye ligand (e.g., FlAsH, ReAsH) and a short genetically encoded tetracysteine (Cys4) peptide tag could be used to label proteins in cells (Figure 1a). Other strategies that have been developed based off of this concept include:
- Self-labeling tags (e.g., SNAP-tag, HaloTag, TMP-tag) – These widely used systems consist of a genetically encoded enzyme variant tag that reacts specifically and irreversibly with a small substrate ligand motif attached to a fluorophore (Figure 1b).
- Engineered ligases (e.g., lipoic acid ligase, biotin ligase, phosphopantetheinyltransferase) – These enzymes catalyze the covalent attachment of a fluorophore ligand to a peptide tag (Figure 1c).
- Click chemistry (e.g., transcyclooctene–tetrazine)– Nonnatural amino acids can be incorporated into a protein structure and then used with the growing toolbox of bioorthogonal chemical reactions (i.e., click chemistry) to attach a fluorophore at the specific site of incorporation (Figure 1d).
- Fluorogen activating proteins (FAPs) – These modified antibody fragments bind and enhance small molecule fluorogens (Figure 1e).
- Stains – These labels consist of a fluorophores conjugated to molecular species with high affinity for endogenous molecular targets such as paclitaxel. Unlike other systems, no genetically encoded tag is required as the small-molecule binding motif targets endogenous protein (Figure 1f).
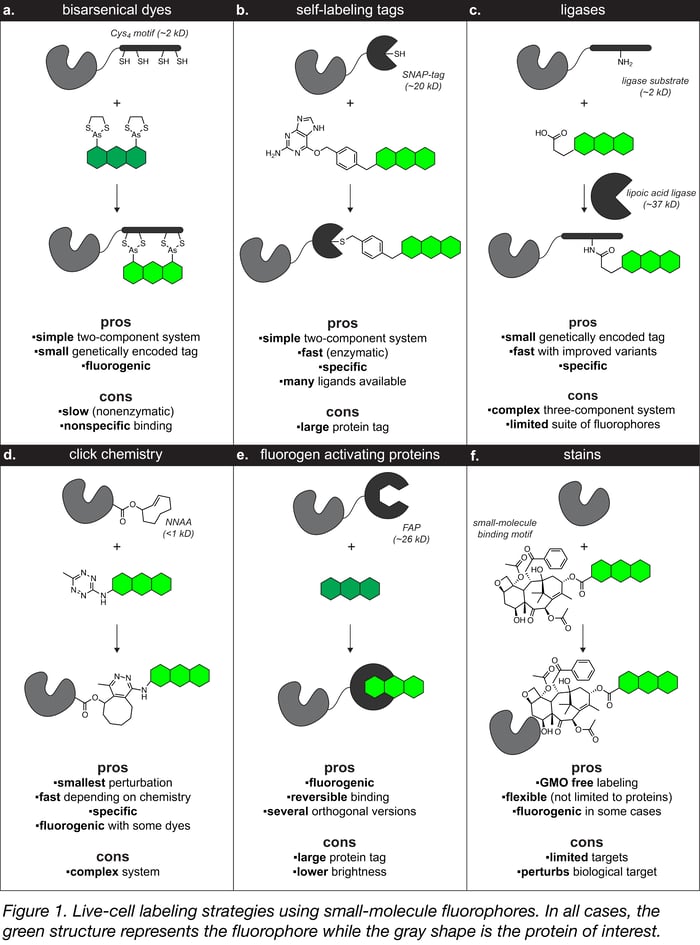
Labeling strategy pros and cons
All of these labeling strategies have trade-offs between the size of the genetically encoded tag, the speed and selectivity of the fluorophore attachment, the brightness of the resulting conjugate, and the complexity of the system. A particularly desirable property for small-molecule labeling strategies is fluorogenicity, meaning the ligand exhibits low fluorescence when free in solution, but high fluorescence upon binding to its cognate protein. In some cases this property eliminates the need for removing excess dye from the sample, which is particularly important for samples where washing is difficult, such as intact tissue. Some chemical labeling strategies such as the bisarsenical dye (Figure 1a) and FAP systems (Figure 1e) are inherently fluorogenic and several others can be made fluorogenic using environmentally sensitive fluorophores. Overall, self-labeling tag systems (Figure 1b) are perhaps the best method for live cell labeling and the easiest switch from fluorescent proteins given the relative simplicity of the system and the availability of fluorescent and fluorogenic ligands.
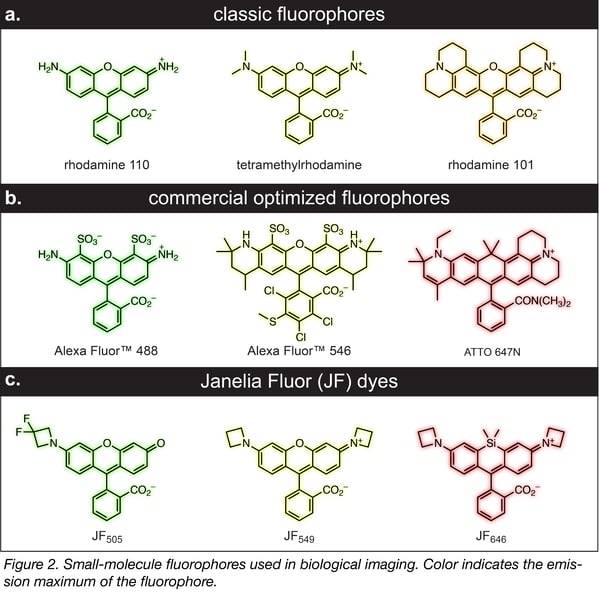
New fluorophores
As a corollary to these innovative labeling strategies, several groups—including mine—have been revisiting the old chemistry of dyes. The first synthetic dye, mauvine, was discovered in 1856 by William Perkin. His discovery set off a flurry of activity and the majority of the classic small molecule fluorophores, such as rhodamines (Figure 2a), were discovered in the 19th century. Many of the small-molecule labeling techniques have focused on this classic, net-neutral dye scaffold due to its established chemistry, small size, brightness, and cell permeability. In past decades, further refinements to this established dye structure have yielded commercial panels of advanced rhodamine fluorophores, such as the Alexa Fluor and ATTO dyes, with improved brightness, photostability, and spectral range. Nevertheless, these fluorophores were designed as antibody and oligonucleotide labels for fixed-cell imaging, not for live-cell applications. Thus, these fluorophores are often bulky and contain polar groups (Figure 2b), which can preclude their use inside living cells. We recently discovered that incorporation of four-membered azetidine rings could substantially enhance the brightness and photostability of the classic rhodamines and their analogs without sacrificing their small size and membrane permeability (Figure 2c). These cell-permeable Janelia Fluor (JF) dyes have excellent properties inside living cells, especially for advanced imaging experiments. We continue to develop derivatives that have different spectral properties, exhibit high fluorogenicity, are photoactivatable, and function in vivo.*
Final thoughts
The ease of use and continual improvement of fluorescent proteins makes them the go-to choice for many (if not most) fluorescence microscopy experiments. However, when your budget gets a bit thin, small-molecule labeling approaches can provide a fresh infusion of photons for your imaging experiment. Innovative labeling strategies and improved fluorophores are making chemical dyes increasingly attractive and accessible to cell biologists—and we are not done yet. Further refinements to these systems—smaller tags, brighter conjugates, higher fluorogenicity, photoactivation etc.—can further increase the photon budget and allow us to push further the frontiers of biological imaging.
*Worried about your budget? Email janeliafluor@janelia.hhmi.org to try out the JF dyes.
Acknowledgements
I thank Brett Mensh, a wordsmith soldier of fortune, for helpful comments.
Many thanks to our guest blogger, Luke D. Lavis! All images in this blog post were contributed by Luke D. Lavis.
 Luke D. Lavis is currently a Group Leader at the Janelia Research Campus, Howard Hughes Medical Institute. He would rather be in the lab making new fluorophores for advanced imaging modalities. Follow him on twitter @rhodamine110.
Luke D. Lavis is currently a Group Leader at the Janelia Research Campus, Howard Hughes Medical Institute. He would rather be in the lab making new fluorophores for advanced imaging modalities. Follow him on twitter @rhodamine110.
References
1. Liu, Zhe, Luke D. Lavis, and Eric Betzig. "Imaging live-cell dynamics and structure at the single-molecule level." Molecular cell 58.4 (2015): 644-659. PubMed PMID: 26000849.
2. Grimm, Jonathan B., et al. "A general method to improve fluorophores for live-cell and single-molecule microscopy." Nature methods 12.3 (2015): 244-250. PubMed PMID: 25599551. PubMed Central PMCID: PMC4344395.
3. Lavis, Luke D., and Ronald T. Raines. "Bright ideas for chemical biology."ACS chemical biology 3.3 (2008): 142-155. PubMed PMID: 18355003. PubMed Central PMCID: PMC2802578.
4. Xue, Lin, et al. "Imaging and manipulating proteins in live cells through covalent labeling." Nature chemical biology 11.12 (2015): 917-923. PubMed PMID: 26575238.
5. Crivat, Georgeta, and Justin W. Taraska. "Imaging proteins inside cells with fluorescent tags." Trends in biotechnology 30.1 (2012): 8-16. PubMed PMID: 21924508. PubMed Central PMCID: PMC3246539.
Additional Resources on the Addgene Blog
- Learn About Other Protein Tags
- Catch Up on Your Fluorescent Protein Background
- Which Fluorescent Protein Should I Use?
Resources on Addgene.org
- Generate Fluorescent Protein Fusions
- Request Plasmids from the Michael Davidson Collection
- Find Fluorescent Protein Kits
Topics: Fluorescent Proteins, Fluorescent Imaging, Non-protein Fluorophores







Leave a Comment