This post was contributed by guest bloggers Keith Pardee and Alexander A. Green.
Zika background
 First identified in 1947 in Uganda, the Zika virus had received little attention and, for the most part, had been associated with low morbidity and mild symptoms. This changed in January with the report of an outbreak of the virus in Brazil that was correlated with greater rates of infection and rare, but severe, symptoms, including the development of fetal microcephaly and Guillain-Barré syndrome. In response, the World Health Organization has declared a global health emergency and called for the fast-tracked development of diagnostics. Mostly transmitted by Aedes mosquitoes (aegypti and albopictus) and, aided by international travel, the Zika virus is expected to expand into heavily populated regions of South, Central, and North America. Diagnostics will play an important role in helping to monitor and slow this spread until vaccine programs can be put in place to provide community protection.
First identified in 1947 in Uganda, the Zika virus had received little attention and, for the most part, had been associated with low morbidity and mild symptoms. This changed in January with the report of an outbreak of the virus in Brazil that was correlated with greater rates of infection and rare, but severe, symptoms, including the development of fetal microcephaly and Guillain-Barré syndrome. In response, the World Health Organization has declared a global health emergency and called for the fast-tracked development of diagnostics. Mostly transmitted by Aedes mosquitoes (aegypti and albopictus) and, aided by international travel, the Zika virus is expected to expand into heavily populated regions of South, Central, and North America. Diagnostics will play an important role in helping to monitor and slow this spread until vaccine programs can be put in place to provide community protection.
Toehold switches for pathogen detection
In 2014, as postdoctoral fellows with Jim Collins and Peng Yin at the Wyss Institute (Harvard University), we developed two new enabling technologies for the field of synthetic biology. The first was a new class of programmable riboregulators, called toehold switches, which allow for the rational design of RNA sensors that can detect virtually any RNA sequence (Green et al, 2014). The second was an in vitro paper-based platform that allowed gene circuits to be deployed out of the laboratory in a cell-free and biosafe format that was stable for up to a year at room temperature (Pardee et al, 2014). As a demonstration of the diagnostic potential of these combined technologies, we built paper-based synthetic gene networks for the detection of antibiotic resistance and strain-specific diagnostics for the Ebola virus. Our original proof-of-concept work was exciting because it enabled gene circuit-based sensors to be used outside of the laboratory. The detection threshold for these RNA sequences (3 nM), however, was quite high and above what would be expected in patient samples (e.g.: blood, urine).
Adapting toehold switches for Zika detection
We and other members of the lab had been working on advancing the paper-based platform toward a more field-ready format when the Zika virus outbreak was reported, and we felt that, given the critical need for low-cost, field-based diagnostics, our approach could make an impact. Our motivation was underscored by the potential scale of the emerging outbreak, as many as 2.2 billion people globally live in areas conducive to the spread of the virus by the Aedes mosquito (Messina et al, 2016). We quickly assembled a team of students, postdocs and professors to move the project forward as quickly as possible and submitted our manuscript to the journal Cell just six weeks later.
The Zika publication timeline
I think we can speak for all the authors on the paper when we say the timeline to develop our paper-based platform into clinically relevant diagnostics for the Zika virus was daunting. It took a true team effort and many late nights, not unlike most research, but the urgent need for the test did catalyze our resolve to get the work done as quickly as we could. From the outset, we spread out the risk for the project by initially designing and ordering several dozen versions of the toehold switches and amplification primers. This helped make sure that we had a number of successful sensors to draw on as our assays grew more stringent, and it avoided any days of lost productivity waiting for follow-up DNA oligos to arrive. We also were careful to set up clear tasks for each member of the team to accomplish so that many elements of the project were able to move forward quickly in parallel. More broadly, we drew on the combined creativity and expertise of the group and yet never hesitated to reach out to the greater research community when we needed additional expertise. Perhaps most striking is that there were seven institutions involved in the effort. This inclusive and open approach by all involved definitely contributed to our ability to complete the project on such a short timescale.
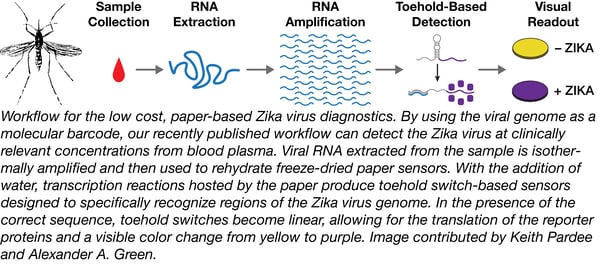
Our article ultimately reported the development of a rapid design and prototyping pipeline that can take pathogen sequence information from public databases to the production of validated sensors in as little as one week (Pardee et al, 2016). Using toehold switch-based RNA sensors, our approach uses portions of the pathogen’s genome as unique barcodes to identify samples containing the virus. Of the 48 sensors, targeting 24 regions of the genome, we have deposited the best performing sensors and constructs encoding corresponding portions of the Zika virus genome with Addgene to facilitate rapid sharing of these tools with the Zika virus research community.
Find the Zika Sensor Constructs at Addgene
As we report, we have embedded this next generation of toehold switch-based RNA sensors within an extensive infrastructure of molecular and electronic companion technologies that has transformed the assay’s capabilities. Specifically this includes primer directed isothermal RNA amplification, new toehold switch designs, CRISPR/Cas9-mediated genotyping, as well as our electronic reader that provides portable quantification of diagnostic results. Our approach now detects samples at clinically relevant concentrations (1-3 fM; 1x106-fold lower than before), and our combined biomolecular tools can distinguish between global strains of the Zika virus (Asian-, African- and American-lineages). Strain discrimination is especially important in the current outbreak because of the neurological symptoms associated with the American-lineage of the virus. For the first time, we have also demonstrated our technology on active virus, as well as shown that we can detect the virus in a clinically relevant setting. Molecular diagnostics for the Zika virus predominantly look at viral load in patient blood and urine samples; we showed that we can detect the virus directly from infected monkey blood plasma without purification or concentration of the virus.
Future work
Going forward, our short-term efforts are focused on finding the resources necessary to advance the technology through product development, the regulatory process, and manufacturing so that we can get the diagnostics into the hands of global health responders as soon as possible. Our longer-term aim is to advance our programmable approach to other critical needs and to further develop our rapid prototyping and sensor assembly pipeline for future emerging pathogens or health crises.
Many thanks to our Guest Bloggers Keith Pardee and Alexander Green!
 Keith Pardee is an Assistant professor in the Leslie Dan Faculty of Pharmacy at the University of Toronto. His research is focused on developing cell-free synthetic biology applications for human health and the environment.
Keith Pardee is an Assistant professor in the Leslie Dan Faculty of Pharmacy at the University of Toronto. His research is focused on developing cell-free synthetic biology applications for human health and the environment.
 Alexander A. Green is an Assistant professor at the Biodesign Center for Molecular Design and Biomimetics and the School of Molecular Sciences at Arizona State University. His group carries out research in synthetic biology, nanomaterials science, and self-assembly for applications in health, energy, and sustainability.
Alexander A. Green is an Assistant professor at the Biodesign Center for Molecular Design and Biomimetics and the School of Molecular Sciences at Arizona State University. His group carries out research in synthetic biology, nanomaterials science, and self-assembly for applications in health, energy, and sustainability.
References
1. Pardee, Keith, et al. "Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components." Cell (2016). PubMed PMID: 27160350.
2. Green, Alexander A., et al. "Toehold switches: de-novo-designed regulators of gene expression." Cell 159.4 (2014): 925-939. PubMed PMID: 25417166. PubMed Central PMCID: PMC4265554.
3. Pardee, Keith, et al. "Paper-based synthetic gene networks." Cell 159.4 (2014): 940-954. PubMed PMID: 25417167. PubMed Central PMCID: PMC4243060.
4. Messina, Jane P., et al. "Mapping global environmental suitability for Zika virus." Elife 5 (2016): e15272. PubMed PMID: 27090089.
Resources on the Addgene Blog
- Read other Synthetic Biology Posts
- Listen to Our Interview with Previous Collin's Lab Member and Sample 6 Cofounder Michael Koeris
- Learn How Minigenomes Are Being Used to Study Dangerous Viruses
Resources on Addgene.org
Topics: Synthetic Biology, Other Plasmid Tools, Plasmids






Leave a Comment