 In this quarterly blog series, we’ll highlight a few of the new CRISPR plasmids available at Addgene. We will still periodically focus on specific CRISPR plasmid tools more in-depth, but we hope that this blog series will help you find more new CRISPR tools for your research!
In this quarterly blog series, we’ll highlight a few of the new CRISPR plasmids available at Addgene. We will still periodically focus on specific CRISPR plasmid tools more in-depth, but we hope that this blog series will help you find more new CRISPR tools for your research!
This time:
- Optogenetic repression in yeast
- Base editing
- CRISPRi in Caulobacter crescentus
- Multiplexed gRNA for human pluripotent stem cells
- CRISPR-based DNA demethylation
Optogenetic repressors of gene expression in yeast
Megan McClean’s lab generated an optogenetic toolkit that controls repression of gene expression in Saccharomyces cerevisiae. The system consists of dCas9, the mammalian transcriptional repressor Mxi1, and an optogenetic tool that controls nuclear localization (LINuS). Blue light renders the NLS accessible and the complex enters the nucleus where dCas9-Mxi1 can repress gene expression. In the dark, the NLS is inaccessible and the complex remains in the cytoplasm.
- Read the paper in Cellular and Molecular Bioengineering
- Find the optogenetic repressor plasmids
Cytosine base editors with minimized off-target DNA editing
The original cytosine base editor, BE3, induces a low frequency of genome-wide off-target DNA editing. Using several rapid and cost-effective screening methods, David Liu’s lab identified and validated CBEs with reduced Cas9-independent deamination. They generated these editors using the BE4max architecture. The new editors reported in this study have ~10-100-fold lower average Cas9-independent off-target DNA editing, ~5–50-fold lower levels of Cas9-dependent off-target DNA editing, and maintain ~50–90% of average on-target DNA-editing levels relative to BE4max.
- Read the paper in Nature Biotechnology
- Find the base editor plasmids
CRISPRi in Caulobacter crescentus
 |
| Figure 1: Serial dilutions of CRISPRi in Caulobacter cells for ctrA gene expression using various dCas9. Image adapted from Guzzo et al., 2020. |
Caulobacter crescentus is a popular model organism for studying the bacterial cell cycle, asymmetric cell division, and cellular differentiation, but it takes several weeks to delete or deplete individual genes. Michael Laub’s lab developed a CRISPRi system to downregulate expression of specific Caulobacter genes using a catalytically dead Cas9 (dCas9) derived from Streptococcus pasteurianus or the type II CRISPR3 module of Streptococcus thermophilus (the popular Streptococcus pyogenes system does not work well in Caulobacter). To test their CRISPRi system, the Laub lab repressed ctrA, a gene encoding an essential cell cycle regulator. The lab expressed both dCas9 and the ctrA sgRNA and saw that the levels of ctrA mRNA decreased after 20 minutes in xylose (which controls expression of dCas9). The lab demonstrated the system can also be used to downregulate multiple genes at once.
- Read the paper in mBio
- Find the CRISPRi plasmids
A multiplexed gRNA piggyBac transposon system for human pluripotent stem cells
Lindy Barrett’s lab developed new fluorescently labeled piggyBac vectors to deliver uniform and sustained expression of multiplexed gRNAs in human pluripotent stem cells. Previously, CRISPRi or CRISPRa depended on continued expression of dCas9 and gRNAs whose expression varies depending on transgene design and delivery. They generated hPSC lines with AAVS1-integrated, inducible, and fluorescent dCas9-KRAB and dCas9-VPR for quantifying and tracking cells that express both the dCas9 construct and the fluorescently-labeled gRNA multiplex.
- Read the paper in Scientific Reports
- Find the plasmids
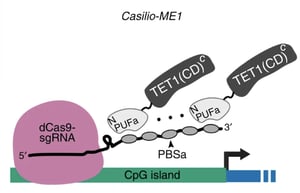
Enhanced CRISPR-based DNA demethylation
|
|
|
Figure 2: The components of Casilio-ME1. Image from Taghbalout et al., 2019. |
Fusing epigenetic modifiers such as TET1 with inactive Cas enzymes can alter gene expression via acetylation or demethylation. Casilio DNA Methylation Editing (Casilio-ME) is a methylation editing tool from Albert Cheng’s lab that enhances RNA-guided methylcytosine editing by TET1 by co-delivering TET1 and protein factors for linking methylcytosine oxidation to DNA repair, which increases activation of methylation-silenced genes.
- Read the paper in Nature Communications
- Find Casilio-ME plasmids
If you have a new CRISPR tool you’ve recently deposited to Addgene and you’d like it to be included in the next What’s New in CRISPR blog post, please let us know. We also welcome blog posts from guest bloggers. Check out this blog post for more information.
Additional resources on the Addgene blog
- Download the CRISPR 101 eBook
- Find our CRISPR 101 blog posts
- Read our CRISPR cheat sheet
Resources on Addgene.org
- Read our CRISPR guide
- Find CRISPR plasmids by function
- Find recently deposited plasmids
Topics: CRISPR, Other CRISPR Tools






Leave a Comment