Adeno-associated viruses (AAV) are an important biological tool that allow scientists to express a gene of interest in a cell or animal model. AAV plasmids contain several important elements, but among the most important are the inverted terminal repeats (ITRs). ITRs are the elements that make an AAV plasmid an AAV plasmid.
Within the AAV genome, ITRs serve as the origin of replication and are required for replication, packaging, and vector persistence. They work in conjunction with the viral rep (replication) and cap (capsid) proteins, which bind to the ITRs and initiate replication. ITRs are cis-acting elements, which means that they are in the same DNA molecule as the gene that they package. In a transfer plasmid, the transgene is flanked by two ITRs, and the ITRs are retained with the gene in a recombinant AAV (rAAV) vector (Figure 1). Everything else outside the ITRs gets left behind.
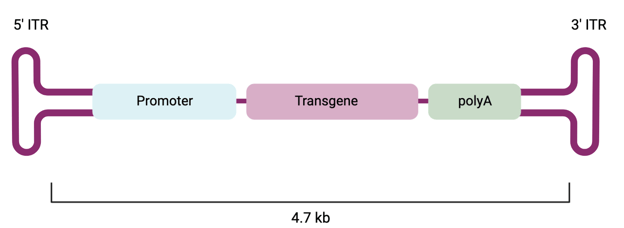
Figure 1: Recombinant AAV vector with ITRs that flank the gene cassette, which contains a promoter, the gene of interest, and a polyA signal. Created with biorender.com.
ITR structure
WT ITRs are symmetrical 145-nucleotide sequences that flank the ends of the single stranded DNA genome of AAV in the native virus. Since the WT sequence is relatively unstable, a more stable 130 bp version is used in most plasmids. In an AAV vector plasmid, they flank the gene of interest. A single ITR consists of palindromic arms (A-A’, B-B’, and C-C’) that give the feature its characteristic T-shape. The arrangement of the B-B’ and C-C’ palindromic sequences determines the orientation of the ITR, which can either be “flip” or “flop” (Figure 2). Additionally, the ITR contains a 4-nucleotide Rep binding element (RBE) that serves as a binding site for Rep78 and Rep68 to initiate replication and a terminal resolution site that serves as the target site for Rep proteins (Savvy et al. 2013).
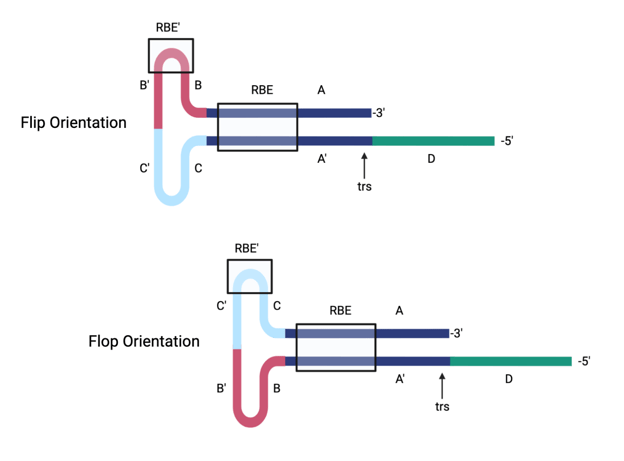
Figure 2: AAV2 wild type ITR in flip and flop orientation. The ITR contains three palindromic sequences (A-A’, B-B’, and C-C’ regions) and forms a T-shaped structure. The Rep binding element (RBE) and terminal resolution site (trs) are noted. Created with biorender.com.
ITR modifications and types
Since ITRs are one of the most important elements of AAV and rAAV vectors, modifications can affect gene transfer efficiency and transgene expression (Zhou et al. 2017). For example, deleting the B-B' or C-C' arms can reduce the packaging efficiency but increase transgene expression (Zhou et al., 2017). The most common variant is an 11 bp deletion in the B or C domain of one ITR. These small deletions are not usually an issue as they are thought to be self-repaired during the AAV replication process (Chen et al., 2004). Mutations in the RBE site reduce binding with Rep proteins and mutations in the trs site reduce Rep nicking. Since Rep proteins are important for viral genome replication, these mutations can affect transduction efficiency and subsequent transgene expression. Replacing the D’ region of an ITR with a sequence containing transcription factor binding sites can increase transgene expression and transduction efficiency (Shitik et al. 2023).
Transgene expression can be altered by using ITR sequences from different AAV serotypes that vary in their RBE and T-shaped hairpin loop sequences (Wilmott et al. 2019). Most ITR sequences are from AAV2 because they demonstrate high promoter activity (Earley et al. 2020), but non-AAV2 ITRs can be used to drive gene expression at varying capacities.
AAV vectors containing the AAV2 ITRs can be cross-packaged with capsids from multiple AAV serotypes (Wilmott et al., 2019), and different combinations of ITR and capsid serotypes vary in their gene transfer efficiency. Our practical knowledge of ITRs is still limited, but this area of research is important for downstream AAV applications.
Tips & FAQs
Due to their high GC content and hairpin structures, ITRs are prone to replication errors and disruptive mutations and truncations that can make vector propagation and sequencing a headache.
How do I maintain ITR integrity and prevent unwanted mutations?
During propagation, mutants rapidly replicate in a growth culture and result in a high percentage of plasmid DNA with mutated ITR sequences. Using recombinant-deficient bacterial strains such as NEB Stable, Stbl3, JC8111, SURE2, or XL10-Gold will help to reduce the likelihood of ITR deletions occurring. Check out our blog post for additional tips to reduce the risk of plasmid recombination.
How do I validate my ITR sequences?
Next-generation sequencing and Sanger sequencing might fail to correctly read through ITR sequences. GC tails are often located immediately outside of the ITRs in the plasmid backbone and do not properly assemble, but these sequences can sometimes be removed to improve vector stability and sequencing results. Next-generation sequencing is also helpful in quantifying ITR mutants since analysis of individual reads allows for identification of non-dominant ITR sequences.
Restriction digests with the enzymes SmaI and SrfI can also be used to validate ITR sequences as these enzymes cut at known sites within the ITR. However, small deletions and point mutations will go undetected by restriction digest methods.
Resources and references
More resources on the Addgene blog
Viral Vectors 101: Viral Vector Elements
Viral Vectors 101: Pseudotyping
Viral Vectors 101: Systemic Capsids
More resource on Addgene.org
Addgene's AVV guide
AAV Viral Preps
References
Chen, Y., Hu, S., Lee, W., Walsh, N., Iozza, K., Huang, N., Preston, G., Drouin, L. M., Jia, N., Deng, J., Hebben, M., & Liao, J. (2024). A Comprehensive Study of the Effects by Sequence Truncation within Inverted Terminal Repeats (ITRs) on the Productivity, Genome Packaging, and Potency of AAV Vectors. Microorganisms, 12(2), 310. https://doi.org/10.3390/microorganisms1202031038399714. PMID: 38399714.
Earley, L. F., Conatser, L. M., Lue, V. M., Dobbins, A. L., Li, C., Hirsch, M. L., & Samulski, R. J. (2020). Adeno-Associated Virus Serotype-Specific Inverted Terminal Repeat Sequence Role in Vector Transgene Expression. Hum Gene Ther, 31(3–4), 151–162. https://doi.org/10.1089/hum.2019.274. PMID: 31914802.
Savy, A., Dickx, Y., Nauwynck, L., Bonnin, D., Merten, O.-W., & Galibert, L. (2017). Impact of Inverted Terminal Repeat Integrity on rAAV8 Production Using the Baculovirus/Sf9 Cells System. Hum Gene Ther Methods, 28(5), 277–289. https://doi.org/10.1089/hgtb.2016.133. PMID: 28967288.
Shitik, E. M., Shalik, I. K., & Yudkin, D. V. (2023). AAV- based vector improvements unrelated to capsid protein modification. Front Med, 10, 1106085. https://doi.org/10.3389/fmed.2023.1106085. PMID: 36817775.
Wilmott, P., Lisowski, L., Alexander, I. E., & Logan, G. J. (2019). A User’s Guide to the Inverted Terminal Repeats of Adeno-Associated Virus. Hum Gene Ther Methods, 30(6), 206–213. https://doi.org/10.1089/hgtb.2019.276. PMID: 31752513.
Zhou, Q., Tian, W., Liu, C., Lian, Z., Dong, X., & Wu, X. (2017). Deletion of the B-B’ and C-C’ regions of inverted terminal repeats reduces rAAV productivity but increases transgene expression. Sci Rep, 7(1), 5432. https://doi.org/10.1038/s41598-017-04054-4. PMID: 28710345.
Topics:
Viral Vectors,
Viral Vectors 101









Leave a Comment