This post was contributed by Adriana Galvan, an associate professor at Emory University School of Medicine.
Optogenetics and chemogenetics are powerful tools to modulate the activity of neurons and other brain cells. Since the opsins or chemogenetic receptors used in these techniques cannot be directly visualized with fluorescence or immunohistochemistry methods, these proteins are routinely fused to ‘tag’ or ‘label’ proteins to facilitate its detection.
Fluorescent proteins are most widely used tags because they emit endogenous fluorescence that allow easy detection with the appropriate fluorescent illumination in in vivo, ex vivo, or post-mortem experiments. Fluorescent proteins can also be identified using antibodies and immunohistochemistry techniques. Thus, these proteins provide a versatile method to identify the expression of transgenes of interest. Furthermore, fluorophore variants (for example green and red fluorescent proteins) allow simultaneous identification of more than one protein of interest.
However, in our experience, we found that fluorescent proteins might not always be the best option.
Fluorescent proteins can interfere with fused protein function
Under some circumstances, the fused fluorescent proteins can be a roadblock, rather than an aid, in the expression of the protein of interest. The green fluorescent protein (GFP) and mCherry, two of the most common tags currently used, are approximately 240 amino acids in size. The size of these proteins, or dimerization between them, could affect the properties or trafficking of the fusion protein (Kimple et al., 2013).
We found that the fluorescent proteins can, indeed, interfere with the correct function of the protein of interest. In our studies, we used AAV viral vectors to express hM4Di in neurons of rhesus monkeys or mice (Galvan et al., 2019). The hM4Di receptor is an engineered Gi protein-coupled receptor, one of the Designer Receptor Exclusively Activated by Designer Drugs (DREADDs) (Armbruster et al., 2007).
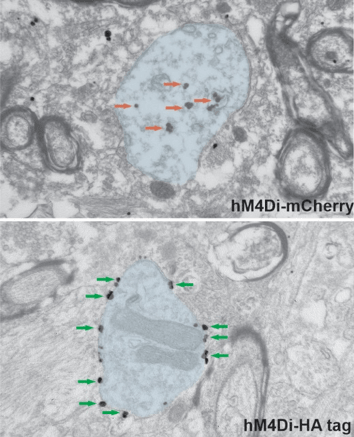
We initially used a construct in which the hM4Di was fused to mCherry. To investigate the localization of the hM4Di, we used primary antibodies against mCherry, and secondary antibodies coupled to nanogold particles, that allow a high-spatial resolution of the protein localization. Surprisingly our observations at the electron microscope showed us that, in monkey neurons, the large majority of the gold particles were in the cytoplasm of dendrites, and only a small proportion were found on the plasma membrane (top panel in Fig. 1). Of course, this was problematic, because the DREADD receptors need to be located on the plasma membrane to be activated by the DREADD actuator after it has been administered. Although we found a larger proportion on the membrane in mice neurons, most of them were still cytoplasmic.
Use of ‘small’ tags to identify proteins
Given the limited expression of hM4Di‐mCherry at the plasma membrane in monkey neurons, we next hypothesized that a different tag fused to the hM4Di might improve the transport of this DREADD to the plasma membrane. The haemagglutinin (HA) tag is a small 9 amino acid epitope of the haemagglutinin of the influenza virus, introduced to the field of fusion proteins more than thirty years ago (Field et al., 1988). Thus, we injected AAVs carrying hM4Di-HA in the brains of mice and monkeys. Surprisingly, we found that this small change made a big difference. Replacing the mCherry with the HA tag resulted in abundant expression of hM4Di in plasma membranes in both species (bottom panel in Fig. 1).
Our results indicate that the tag protein may have an important influence on the expression of the transgene of interest and its localization in the membrane. The lesson we learned from these studies is that the tag protein can impact the expression of the transgene of interest. In our studies, mCherry interfered with the transport to the membrane of the DREADDs. In studies where the fluorescence is not needed, the use of HA and other short epitope tags (Kimple et al., 2013) could help improve the transport of protein of interest the membrane.
 |
|
Figure 1: Electron microscope images showing ultrastructural localization of hM4Di in monkeys. The pre-embedding immungold technique was used to reveal the hM4Di coupled to mCherry (top) or HA tag (bottom). The images show examples of dendrites (blue shades) in the monkey amygdala. Red and green arrows point, respectively, to gold particles localized in the cytoplasm or bound to the plasma membrane. Note that the use of the HA tag significantly improves the transport of hM4Di to the plasma membrane. |
Our studies provided evidence that some fluorescent proteins can be, in some circumstances, detrimental to the expression of the protein of interest. However, we also found that this is not generalizable to all fusion proteins. For example, we saw that another type of DREADD, hM3Dq, is successfully expressed at the plasma membrane, even when fused to mCherry. But our data does highlight the importance of a careful characterization of transgene expression.
Which tag should I use?
When deciding on the appropriate tag, the experimental design should be the main guidance. The results from our studies suggest that non-fluorescent tags may be advantageous over fluorescent proteins when the fluorescence is not essential to identify the expression of the opsin or chemogenetic receptor. However, in some cases the fluorescent signal may be critical for the experiment. Since the fused protein could represent a hindrance to the successful expression of the protein of interest, an alternative could be bicistronic expression of the fluorescent protein with the opsin or chemogenetic receptor, through an IRES or 2A sequence. In this case, the proteins are not fused. The tag protein will be expressed in the same cell, but not necessarily at the same subcellular compartment as the opsin or chemogenetic receptor. This may not be an important limitation in many studies, and could even be beneficial in some types of experiments (for instance, when the fluorescence expressed in the neuropil conceals identification of opsin-expressing cells).
For other things to keep in mind when making fluorescent protein fusions, check out Joachim Goedhart’s article on the Addgene blog: Fluorescent Proteins 101: GFP Fusion Proteins – Making the Right Connection.
 Thank you to our guest blogger!
Thank you to our guest blogger!
Adriana Galvan is an associate professor at Emory University School of Medicine and a researcher at the Yerkes National Primate Research Center. Using a combination of in vivo and anatomical techniques, her research aims to gain a deeper understanding of the physiology and pathology of the basal ganglia in movement disorders.
References and Resources
References
Armbruster BN, Li X, Pausch MH, Herlitze S, Roth BL (2007) Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proceedings of the National Academy of Sciences 104:5163–5168 . https://doi.org/10.1073/pnas.0700293104
Field J, Nikawa J, Broek D, MacDonald B, Rodgers L, Wilson IA, Lerner RA, Wigler M (1988) Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol Cell Biol 8:2159–2165 . https://doi.org/10.1128/mcb.8.5.2159
Galvan A, Raper J, Hu X, Paré J, Bonaventura J, Richie CT, Michaelides M, Mueller SAL, Roseboom PH, Oler JA, Kalin NH, Hall RA, Smith Y (2019) Ultrastructural localization of DREADD s in monkeys. Eur J Neurosci 50:2801–2813 . https://doi.org/10.1111/ejn.14429
Kimple ME, Brill AL, Pasker RL (2013) Overview of Affinity Tags for Protein Purification. Current Protocols in Protein Science 73: . https://doi.org/10.1002/0471140864.ps0909s73
Additional references on the Addgene blog
- Browse Fluorescent 101 blog posts
- Read about chemogenetics vs. optogenetics
- Find more about the many types of protein tags
References on Addgene.org
- Check out Addgene's Chemogenetics Guide
- Find fluorescent proteins at Addgene
Topics: Optogenetics, Viral Vectors, Chemogenetics






Leave a Comment