When cancers are treated with drugs designed to hit them right where it hurts, the effects are often remarkable but fleeting.
“What’s been shown by others is that, in a relatively short amount of time, cancers become resistant to drugs, particularly targeted therapies,” said Kris Wood of Duke University Medical Center. “While we do not yet have a comprehensive understanding of how cancers become resistant, an emerging theme is that they do so by activating signaling pathways controlling properties like growth, survival, and differentiation.”
 https://www.addgene.org/kits/sabatini-wood-cancer-pathway-orfs/
https://www.addgene.org/kits/sabatini-wood-cancer-pathway-orfs/
Finding signaling pathways involved in drug resistance
Wood wanted to find out which particular pathways drive resistance to specific drugs in specific contexts, so he assembled a plasmid kit designed to do exactly that.
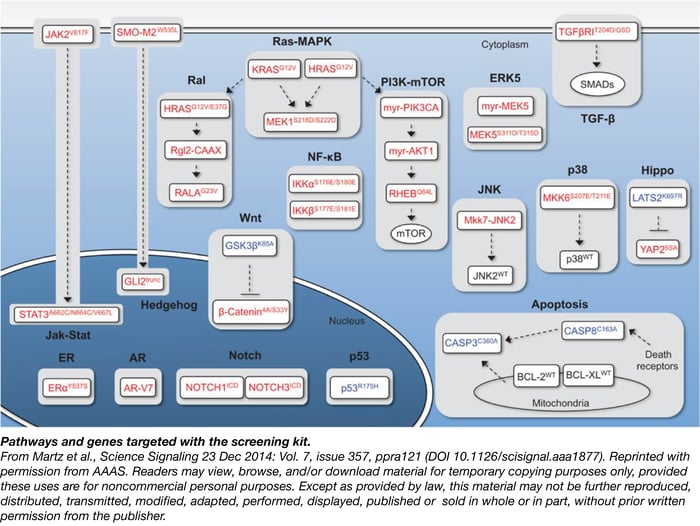
As described in the journal Science Signaling in December 2014, he and his colleagues at Duke University, in collaboration with “cloning guru” Kathleen Ottina and David Sabatini at the Whitehead Institute and MIT, started with a list of 17 signaling pathways that are frequently implicated in cancer cell proliferation, survival, differentiation, and apoptosis. For each of these 17 pathways, they identified a set of one to three mutant complementary DNAs (cDNAs) representing core nodes that, when over-expressed, constitutively activated or inactivated the pathway.
“The idea is that if you introduce them into cells one at a time, you could identify those that confer resistance to drugs,” in a rapid, straightforward, and scalable manner, Wood said.
Thankfully, the researchers didn’t have to discover new ways to activate each of these pathways. Rather, they relied on published studies in the literature to curate the perfect library, obtained the appropriate cDNAs from other researchers, and cloned them into lentiviral vectors to generate a kit which is now available at Addgene.
“We called it cloning by mail,” Wood says, and it took them about six months from start to finish. They cloned all the cDNAs into a single consensus lentiviral expression vector and attached a DNA barcode to each to make the library usable for pooled screens.
Applications of the cancer pathways ORFs kit
Wood and his colleagues developed the kit for use in screening for pathways that drive resistance to drugs in vitro, and they’ve shown that it works. But, he says that’s not the only way to use the kit.
“You could use it to screen for pathways that confer any phenotype of interest, not just drug resistance,” he said. “It could [be used to screen for] morphological features, migration, or survival in different contexts.”
It’s also possible you could make use of these plasmids in living animals, to explore pathways that confer metastatic potential or the ability to survive in certain environments, for example.
“We’ve been contacted by investigators all over the world who are interested to use our libraries to do things - sometimes drug screens and sometimes totally different experiments we never thought of - and that’s thrilling to us,” Wood says. “We want to make these available as widely as possible.”
The researchers have plans to expand the library to include activators for more cancer-related pathways and more nodes within those pathways. With support from the National Cancer Institute, they expect to add another 200 or 250 constructs to the kit in the next two years.
References
1. Martz, Colin A., et al. "Systematic identification of signaling pathways with potential to confer anticancer drug resistance." Science signaling 7.357 (2014): ra121-ra121. PubMed PMID: 25538079. PubMed Central PMCID: PMC4353587.
Additional Resources on the Addgene Blog
- Learn How the Clon Tracer Library Has Been Used in Cancer Lineage Tracing
- Read How CRISPR Can Be Used in Genome-Wide Screening
- Browse 28 Hot Plasmid Technologies from 2015
Additional Resources on Addgene.org
- Find Other Kits for Your Research
- Find Pooled Libraries for Your Research
- View our Hot Plasmid Pages to See the Latest Plasmid Technologies
Topics: Cancer, Other Plasmid Tools, Plasmids






Leave a Comment