One way to define a protein’s purpose is by its protein-protein interactions (PPIs). These interactions are often modeled as binary relationships, i.e. protein A interacts with protein B; but proteins are social biomolecules. They can be part of multiple dynamic and overlapping complexes that have distinct functions. Many existing methods for identifying PPIs, such as affinity purification mass spectrometry (AP-MS), lack the ability to specifically identify proteins that interact with a particular protein complex as opposed to an individual protein. The Bethune Lab has overcome this limitation by creating Split-BioID, a spatiotemporally controllable version of the proximity-dependent biotinylation technique BioID. The key advantage of Split-BioID is that it allows for the validation of a binary PPI as well as the identification of additional interacting factors.
Problems with Existing Methods for Studying Protein-Protein Interactions
- Difficult to detect weak and/or transient PPIs. Many PPI identification techniques rely on protein interactors to stay in close contact with the bait protein throughout a pulldown step, such as AP-MS. This requirement often prevents the identification of weak and/or transient interacting proteins.
- No spatiotemporal control. Many PPI detection methods lack spatiotemporal control or an “on/off switch.” This makes them great tools for providing a global overview of potential interactors with a bait protein but the flip side is that it’s difficult to know which proteins interact with the bait at a particular point in time, i.e. when the bait is interacting with a particular protein partner or when the bait is part of a particular protein complex.
- Not performed in a native cell context. For AP-MS, cells are lysed before the bait protein is pulled down, typically with an antibody. This can artificially select against PPIs that aren't stable outside the native cell context.
The Split-BioID method
Enter Split-BioID, a spatiotemporally controllable version of the BioID protocol. The critical component for both BioID and Split-BioID is BirA*, a promiscuous version of BirA, an E. coli biotin ligase. In BioID, BirA* is fused to a bait protein and it biotinylates proteins that are within a ~10 nm radius of the bait. In Split-BioID, BirA* is first split into two non-functional fragments. When these two fragments are near each other, they combine forces to form a functional BirA* enzyme. This type of experiment is called a protein fragmentation complementation assay (PCA).
In the Split-BioID method, each BirA* fragment is then fused to one of two bait proteins that are known to interact. When this interaction occurs, BirA* activity is restored, and nearby proteins are labeled with biotin. For labeling, BirA* fusion proteins are expressed in cells via plasmids or by the creation of stable cell lines and these cells are then grown in biotin-containing media.
FRET is another example of a protein fragmentation complementation assay that can be used to study binary protein interactions. However, FRET generally only tells you if two proteins interact and cannot be used to identify interaction partners other than those which you’ve already fluorescently labeled.
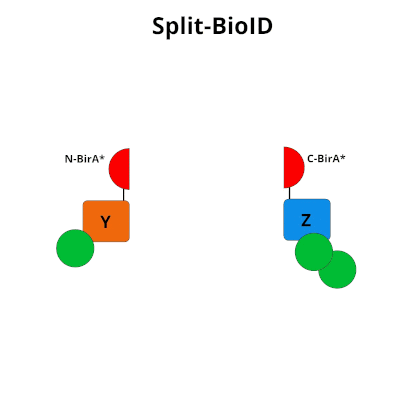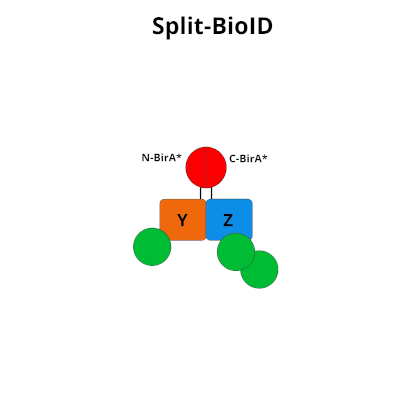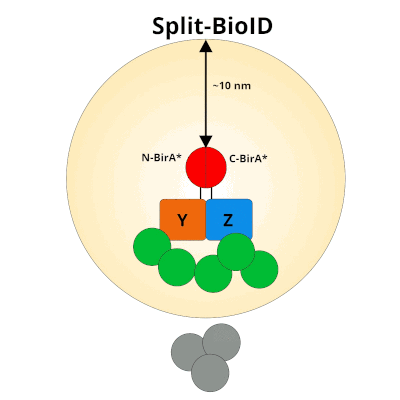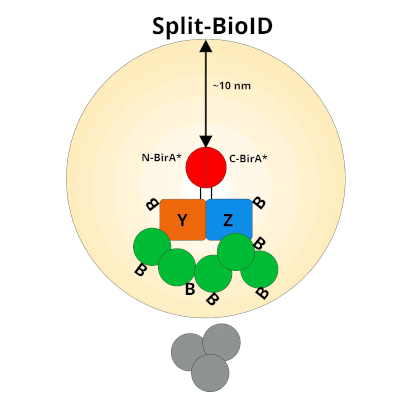
Figure 1: Using Split-BioID to Identify Protein Protein Interactions. Split-BioID is a protein complementation assay where BirA*, an E. coli biotin ligase is split into two non-functional fragments: N-terminal BirA* (N-BirA*) and C-terminal BiraA* (C-BirA*). These two fragments are fused to two proteins, here proteins Y and Z, that are known to interact. When proteins Y and Z interact, the two BirA* fragments complement each other to form a functional BirA* enzyme. When this interaction occurs, BirA* activity is restored, and nearby proteins are labeled with biotin. When proteins Y and Z part ways, BirA* biotinylation activity ceases.
After biotin tagging of proteins in both BioID or Split-BioID, cells are lysed, protein extracted, and labeled protein interactors purified via streptavidin pulldown. Following trypsin digest, PPIs are identified via mass spec analysis. Biotinylated proteins are considered possible interaction partners if they are enriched relative to controls using Split-BioID-GFP and BioID run on six unrelated proteins.
One caveat of Split-BioID is that the orientation of the BirA* fusion matters! Schopp et al saw different levels of fusion protein expression and biotinylation depending on which terminus of the bait proteins were tagged but also which fragment of BirA*, either the N- or the C-terminus, the bait proteins were tagged with. It’s best to test all combinations and pick the conditions that give the strongest biotinylation signal, as measured via Western blot, and that result in proper localization of the fusion protein.
Advantages of Split-BioID
Split-BioID’s key advantage over other PPI methods, including BioID, is its spatiotemporal controlled labeling of interacting proteins; nearby proteins are biotinylated when and where the two bait proteins interact, but labeling ceases when these two bait proteins stop interacting. This feature is particularly useful for in-depth characterization of dynamic protein complexes.
Split-BioID, like BioID, is also useful for identifying weak and/or transient protein-protein interactions since the biotin labeling remains even if the interaction with the BirA* fused bait proteins ceases. Lastly, Split-BioID and BioID identify proteins that interact with the bait in a native cell contact. Other PPI methods may require protein interactions to remain stable (or to be stabilized) during cell lysis and lysate fractionation.
Important considerations for Split-BioID
It’s important to remember that with Split-BioID, and BioID, any protein that’s within a ~10nm neighborhood of the bait protein will be biotinylated. This makes it’s impossible to distinguish proteins that directly interact with the bait from those that indirectly interact or from proteins that are just in the proximity of the bait proteins. Instead of pinpointing specific protein-protein interactions, SplitBioID provides a candidate list of proteins that are part of the bait proteins’ interactome. Further characterization of these candidates is critical to understanding how they interact with the bait proteins.
Another potential limitation of Split-BioID is its speed of labeling. BioID requires 6-24 hours for tetracycline induced expression of the BirA* fusion protein and saturation of biotinylation signal. For Split-BioID, Schopp et al allowed 24 hours for tetracycline induced expression and biotinylation of interacting proteins. This extended incubation period could make it difficult to detect proteins with short half-lives.
Interested in using Split-BioID with your research? Plasmids are available from Addgene!
References
Schopp, I.M., Ramirez, C.C., Debeljak, J., Kreibich, E., Skribbe, M., Wild, K., & Béthune, J. (2017). Split-BioID a conditional proteomics approach to monitor the composition of spatiotemporally defined protein complexes. Nature communications. PubMed PMID: 28585547. PubMed Central PMCID: 5467174.
Munter, S.D., Görnemann, J., Derua, R., Lesage, B., Qian, J., Heroes, E., Waelkens, E., Eynde, A.V., Beullens, M., & Bollen, M. (2017). Split-BioID: a proximity biotinylation assay for dimerization-dependent protein interactions. FEBS letters, 591 2, 415-424. PubMed PMID: 28032891.
- This is a different version of Split-BioID. BirA* is fragmented at a different site than in Schopp et al. Find these Split-BioID Plasmids Here!
Burke, B., Kim, D.I., Roux, K.J., & Raida, M. (2012). A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. The Journal of cell biology. PubMed PMID: 22412018. PubMed Central PMCID: 3308701.
Macneill, S., & Varnaitė, R. (2016). Meet the neighbors: Mapping local protein interactomes by proximity‐dependent labeling with BioID. Proteomics. PubMed PMID: 27329485. PubMed Central PMCID: 5053326.
Additional Resources on the Addgene Blog
- Learn about techniques for visualizing protein turnover
- Learn about plasmids for in vivo biotinylation of bacterial fusions proteins
- Study Protein-DNA Interactions with CUT&RUN
Resources at Addgene.org
- Find plasmids from the Bethune Lab
- Find Plasmids from the Roux Lab
- Find Plasmids for FRET

Topics: Other Plasmid Tools, Plasmids





Leave a Comment