What is polymerase chain reaction?
If you have ever worked in a molecular biology laboratory you have likely done a polymerase chain reaction (PCR). PCR is an in vitro method in which a small amount of DNA can be copied many many times in a short time period. PCR was invented in the early 1980s by Kary B. Mullis who later shared a Nobel Prize in Chemistry for his work. Since then PCR has become a standard and essential practice in molecular biology and can be used in a multitude of scientific techniques such as molecular cloning or molecular diagnostics.
Steps of PCR
The beauty of PCR is that it can amplify DNA using only a short list of reagents and several heating and cooling steps. PCR relies on heat resistant DNA polymerase from the thermophilic bacterium, Thermos aquaticus (Taq). Taq polymerase is thus a heat resistant enzyme that can withstand changes in temperature. Taq was first identified in the late 1960s during research at hot springs in Yellowstone National Park.
In addition to Taq DNA polymerase, PCR requires free nucleotides (dNTPs), template DNA to amplify from and unique single stranded DNA primers that bind upstream (5’) and downstream (3’) of the DNA region of interest. Primers are crucial for this process as DNA polymerases require an existing strand of DNA to add nucleotides to.
Using these reagents and a series of heating (denaturing) and cooling (annealing) steps Taq polymerase can copy DNA between the primers using the dNTPs.
Let’s jump into the specifics of the 3 basic steps of a PCR reaction:
- Denaturation- To amplify DNA, the two strands of the template DNA first have to be separated. This occurs by heating the dsDNA template to a point where the hydrogen bonds break between the base pairs. This results in the separation of the two DNA strands.
- Annealing- The temperature is then dropped to a range in which the forward and reverse primers are stable. At this temperature the primers can anneal to the single stranded DNA template strands. DNA polymerase is also stable at this temperature and can bind to the primers.
- Extension- The temperature is then raised slightly to Taq polymerase’s ideal temperature (70-75oC). At this temperature Taq polymerase can synthesize and elongate the target DNA quickly and accurately.
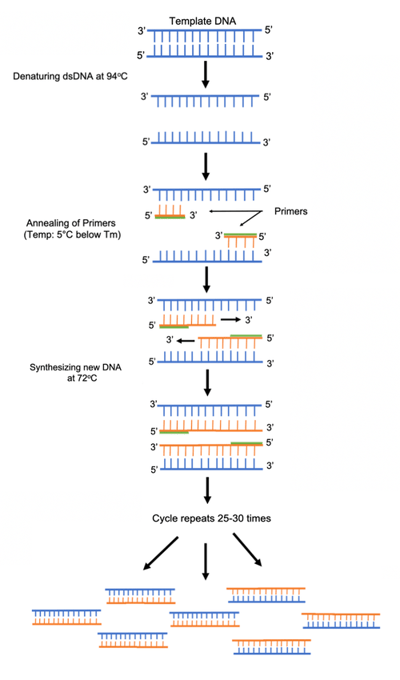 |
| Figure 1: Steps in PCR reaction |
This process of denaturation, annealing and extension is repeated 25-35 times to exponentially replicate the target DNA of interest. This entire reaction used to be done manually in water baths but is now easily done in a programmed thermocycler. For more details on this procedure check out our PCR protocol page, protocol video and reference on how to design PCR primers.
Types of PCR
Since the invention of PCR, different PCR methods have been developed for different scientific applications. These PCR methods all use the same basic PCR set up and steps but differ in how the PCR products are analyzed.
End point PCR
End point PCR, as the name implies, analyzes the end product of PCR temperature cycling. The final PCR product is often visualized on a diagnostic agarose gel to confirm product presence, size, and relative quantity. End point PCR is most commonly used in molecular cloning, sequencing, and genotyping. It’s extremely useful but is not as quantitative as other methods of PCR. Theoretically scientists should be able to determine the quantity of DNA after a PCR reaction as the amplicon doubles every reaction cycle. However, it is common for dNTPs and other reagents to run low during the final cycles which would slow or stop PCR amplification. Other times, the PCR reaction may not be 100% efficient and produce a reduced amount of product.
End point PCR, which is commonly referred to as simply PCR, is used for an array of different molecular biology techniques and experiments. End point PCR’s goal is to amplify a specific target of DNA. This amplicon can be used in molecular cloning to create a plasmid of interest, for detecting the presence of an insert or piece of DNA in a sample such as in colony PCR, or for generating mutated sequences for site-directed mutagenesis.
Quantitative polymerase chain reaction, qPCR
Quantitative polymerase chain reaction (qPCR) also known as real time PCR is a PCR technique used for measuring a starting DNA concentration using PCR. qPCR requires the addition of a probe based fluorescent dye that intercalates with any dsDNA and the use of a fluorometer feature built into the thermocycler to measure that fluorescent output. With this fluorescent dye, the concentration of the DNA during the PCR reaction cycles is continuously detected via a fluorescent signal. The signal increases proportionally to the amount of product produced each cycle.
To determine the concentration of the starting template DNA the fluorescent signal throughout the reaction is compared to a standard curve of amplified DNA of a known starting concentration. The cycle in which the unknown DNA is detected compared to the standard curve can be used to determine the amount of starting material in your sample.
qPCR can also be used to quantitate RNA levels using reverse transcription-qPCR (RT-qPCR). The first step of this process requires RNA to be converted to cDNA using reverse transcription and the cDNA is subsequently quantified by qPCR. qPCR is used in a variety of applications including gene expression profiling, studying copy number variation, and molecular diagnostics.
Digital droplet PCR, ddPCR
Digital droplet PCR or ddPCR is a method that provides ultrasensitive and absolute nucleotide concentration, unlike qPCR where results can vary across replicates. ddPCR can be used to quantify DNA sequences that are rare, for example, rare alleles or mutations. ddPCR also does not require a reference or standard curve, which can be time consuming and challenging to get right. ddPCR uses a water-oil emulsion droplet technology that fractions the PCR reaction sample into approximately 20,000 droplets. Each droplet contains the material required for PCR amplification. Following the PCR reactions each droplet is analyzed by a droplet reader, which measures the fluorescence amplitude of each droplet. The fraction of fluorescent PCR-positive droplets is determined and then analyzed using Poisson statistics to determine the concentration of the original template DNA in the sample.
Addgene currently uses ddPCR for AAV titrating. You can learn all about ddPCR for AAV titration and find helpful tips and tricks in our blog post “Droplet Digital PCR for AAV Quantification.”
Multiplex PCR
Multiplex PCR, as the name implies, is a method in which multiple targets can be amplified in a single PCR experiment using multiple primers all in one PCR reaction. This is an extremely useful PCR method that can help save time and effort in the laboratory.
There are two main categories of multiplex PCR:
-
Single template PCR reaction - one template is amplified using several forward and reverse primer sets.
-
Multiple template PCR - multiple templates with different primer pairs that align to the target region of each template are used in one reaction.
Multiplex PCR is used commonly in disease or pathogen identification. Scientists can simultaneously detect several different pathogens in one specimen saving both time and effort. Scientists can also utilize multiplex qPCR to quantify the concentrations of multiple starting DNA templates in one sample. It is important to note however, that multiplex PCR is more complicated to develop and is often less sensitive than PCRs that use a single pair of primers, like those stated above. Multiplex PCR is also used in high throughput SNP genotyping, gene deletion analysis, and RNA detection.
A summary of the types of PCR
| Type of PCR |
Goal of the PCR method |
Uses of PCR method |
|
End point PCR or just PCR |
Amplify a region of interest |
|
|
qPCR |
Quantify starting template DNA concentration |
|
|
ddPCR |
Determine ultrasensitive and absolute nucleotide concentration of starting material |
|
|
Multiplex PCR |
Amplify multiple targets from one sample |
|
References and Resources
References
-
Shen C-H (2019) Amplification of Nucleic Acids. In: Diagnostic Molecular Biology. Elsevier, pp 215–247
Additional resources on the Addgene blog
- Droplet Digital PCR for AAV Quantification
- Plasmid cloning by PCR
- Plasmids 101: Colony PCR
- Site Directed Mutagenesis by PCR
Resources on Addgene.org
Topics: Molecular Biology Protocols and Tips, PCR






Leave a Comment