Since Addgene began producing AAV vector preparations, we have used quantitative PCR to evaluate the physical titer of our preps. We often repeat this assay to confirm that our results are accurate. But what if we could repeat this assay 10,000 times to ensure that our titers were as accurate as possible? This is where droplet digital PCR (ddPCR) comes in.
AAV titering using quantitative PCR
Before we dive into the details of ddPCR, we should first note that quantitative PCR (qPCR) has been a powerful tool for quantifying AAV for some time now. When titrating AAV by qPCR, viral DNA is amplified and monitored in real time. When the PCR is complete, the results are analyzed by comparing the threshold fluorescence of the viral DNA to the threshold fluorescence of a standard - a set of reactions with known quantities of DNA. Additionally, an AAV reference, a virus with a known titer, can be used to confirm that the standard curve is giving an accurate readout of your samples.
Of course, if qPCR were perfect there would be no need for ddPCR. One issue with qPCR is that the results can vary by a factor of 2. This means that if you set up two identical assays with the same sample, you could end up with a titer of 4 x 1012 genome copies/mL or 8 x 1012 genome copies/mL and both results would be valid. This is why the AAV reference is critical. It allows you to determine if your titers are within the expected range.
We have also found that generating the standard curve for qPCR can be challenging to get right. Here at Addgene, we use a linearized plasmid standard. However, even when the standard is made properly, it is unstable and may only work for a small number of qPCR runs before a new one is needed. For these reasons, we are in the process of transitioning our titering method from qPCR to ddPCR.
Droplet digital PCR (ddPCR) does not require a standard or a reference, meaning saving on reagents (and time)!
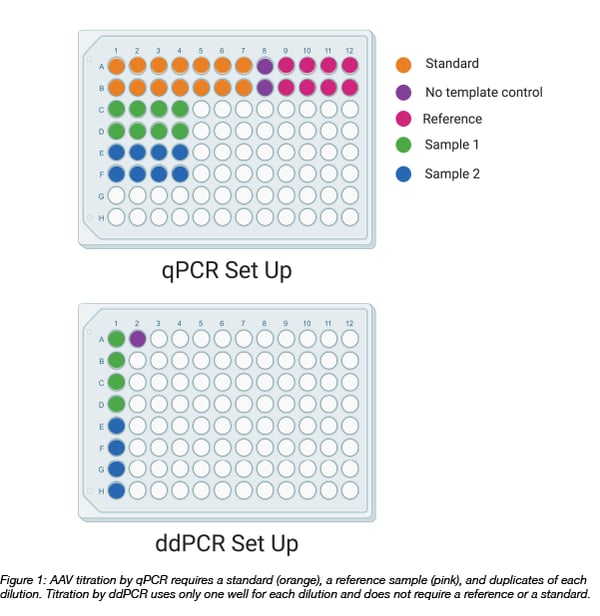
AAV titering using droplet digital PCR
Droplet digital PCR involves partitioning a PCR reaction mixture into approximately 20,000 droplets using water-oil emulsion technology. Each droplet contains the ingredients for amplification of the target DNA. This partitioning reduces the number of PCR inhibitors per reaction and allows for enhanced detection of the product. As an added benefit, replicates are built into the technology. One well (containing thousands of droplets) can be sufficient to capture the information needed for your PCR experiment.
The process to titrate AAV by ddPCR begins with diluting the virus. It is important to note that the dynamic range of the ddPCR is between 1 and 100,000 genome copies (GC) per reaction. Since AAV titers tend to be in the range of 1012 to 1013 GC/mL, you must serially dilute your virus before adding your sample to a mastermix. At Addgene, we typically load 3 dilutions onto our ddPCR plate. As the titer is unknown, each dilution should fall within the range of the assay for a wide range of titers. We usually dilute our samples 1:6 million to 1:25 million.
After making the dilutions, they are transferred to a new plate containing the mastermix which includes primers and a ddPCR supermix. Note that the supermix and droplet oil that you use should come from the same manufacturer. Otherwise you will end up with poor droplet quality.
Using a droplet generator, the sample and the droplet generation oil are moved through small channels to create a water-in-oil emulsion containing approximately 20,000 droplets. Each droplet contains the material required for a mini amplification reaction to take place.
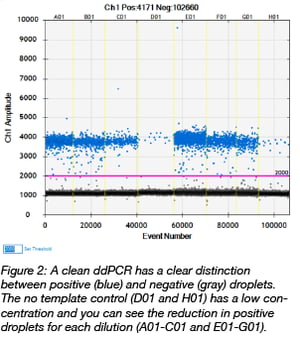 When the PCR is complete, a droplet reader extracts the droplets from the plate and measures the fluorescence amplitude of each one. Droplets that fluoresce contain the amplified target sequence. After all of the droplets have been read, the software outputs an image of the fluorescence amplitude measured for each droplet in each well (Figure 2). A clean ddPCR should have a clear separation between positive (blue) and negative (gray) droplets. The no template control wells should have very few positive droplets. In the image to the right, there is approximately 1 positive copy per microliter in the no template control wells. You’ll also notice a reduction in the number of positive droplets as the dilutions increase.
When the PCR is complete, a droplet reader extracts the droplets from the plate and measures the fluorescence amplitude of each one. Droplets that fluoresce contain the amplified target sequence. After all of the droplets have been read, the software outputs an image of the fluorescence amplitude measured for each droplet in each well (Figure 2). A clean ddPCR should have a clear separation between positive (blue) and negative (gray) droplets. The no template control wells should have very few positive droplets. In the image to the right, there is approximately 1 positive copy per microliter in the no template control wells. You’ll also notice a reduction in the number of positive droplets as the dilutions increase.
The ddPCR software uses the ratio of positive to negative droplets is used to calculate the concentration of the sample. This concentration can then be used to calculate the viral titer:
GC/mL = {[(R*C)(1000/V)]*D}
R = Reaction volume
C = Copies/uL
V = Volume of virus in reaction mix
D = Dilution factor of virus
Most droplet readers have a few channels for detecting fluorescence so that it is possible to measure the concentration of multiple targets simultaneously. For AAV titration, you should only need to use one. However, two channels can be used to examine the integrity of the viral genome (Furuta-Hanawa et al., 2019).
Tips and tricks for performing ddPCR
Clean everything
You must have a clean working area to titer your AAV by ddPCR. Droplet digital PCR is a sensitive assay, so cross contamination into a no template control (NTC) well is common. Here are some steps you can take to achieve a clean NTC:
- Have a dedicated bench with a dedicated set of pipettes for ddPCR set-up.
- Prepare the master mix in a separate area from where you prepare your sample dilutions.
- Aliquot all of your reagents into single-use tubes and grab a fresh one for each set up.
- Wipe down the bench and all consumables with 10% bleach.
Pipette slowly
If you are using a manual droplet generator, you will have to be careful about transferring your droplets from the droplet generator to the PCR plate. Maintaining a high droplet count is important for calculating the concentration of your sample.
Even if you are using an automated droplet generator, you should avoid pipetting too quickly when making your virus dilutions. This will reduce aerosols that could potentially lead to contamination.
Optimize your PCR
For a good starting place on what PCR parameters to use, see Lock’s seminal paper on AAV titration (Lock et al., 2014).
If you are having difficulty getting a clean separation between positive and negative droplets with these parameters, here are a few things you can try:
- Increase the number of cycles. After additional rounds of amplification, your fluorescence amplitude will increase, leading to greater separation of your positive and negative droplets. No more than 50 cycles is recommended.
- Decrease your ramp rate. A rate of 2C/s is recommended to ensure an even temperature change among all of the droplets, but you can go as low as 1C/s.
- Increasing the elongation time to 2 minutes and the denaturation time to 1 minute has been shown to increase droplet separation (Witte et al., 2016). This is particularly important if your amplicon is longer than 150 base pairs.
While we use ddPCR for AAV titration, there are many applications for the technology. Droplet digital PCR is well suited for the detection of low copy numbers. For this reason, it can be used in the detection of rare sequences and single cell analysis. It is also being used in the detection of microbes. Some applications include measuring viable probiotics and detecting circulating pathogens.
With its many applications and ease of use, droplet digital PCR is becoming a popular option for PCR experiments.
References
Furuta-Hanawa, Birei, Teruhide Yamaguchi, and Eriko Uchida. "2D droplet digital PCR as a tool for titration and integrity evaluation of recombinant adeno-associated viral vectors." Human Gene Therapy ja (2019). PubMed PMID: 31140327. PubMed Central PMCID: PMC6707039.
Gobert, Guillaume, et al. "Droplet digital PCR improves absolute quantification of viable lactic acid bacteria in faecal samples." Journal of microbiological methods 148 (2018): 64-73. PubMed PMID: 29548643.
Lock, Martin, et al. "Absolute determination of single-stranded and self-complementary adeno-associated viral vector genome titers by droplet digital PCR." Human gene therapy methods 25.2 (2013): 115-125. PubMed PMID: 24328707. PubMed Central PMCID: PMC3991984.
Song, Neng, et al. "Detection of circulating Mycobacterium tuberculosis-specific DNA by droplet digital PCR for vaccine evaluation in challenged monkeys and TB diagnosis." Emerging microbes & infections 7.1 (2018): 1-9. PubMed PMID: 29691363. PubMed Central PMCID: PMC5915492.
Witte, Anna Kristina, et al. "A systematic investigation of parameters influencing droplet rain in the Listeria monocytogenes prfA assay-reduction of ambiguous results in ddPCR." PloS one 11.12 (2016): e0168179. PubMed PMID: 27992475. PubMed Central PMCID: PMC5167268.
Additional resources on the Addgene blog
- Read more about polymerase chain reaction
- Browse our blog posts related to viral vector protocols and tips
- Read all of our viral vectors blog posts
Resources on Addgene.org
- Learn our protocol for AAV titration by qPCR
- Browse our collection of viral vector related protocols
- Learn more about the Addgene viral service
Topics: Viral Vectors, Viral Vector Protocols and Tips, AAV






Leave a Comment