Site-directed mutagenesis (SDM) is one of the key tools researchers use to prove causation in molecular biology and genetics. It can be used to characterize the function of certain regions in a promoter or gene, as well as to study the effects of inactivating/activating mutations. In biomedical research, modeling patient mutations using SDM can help determine if a variant is causal for a given disease. CRISPR has made genomic SDM relatively straightforward, but plasmid-based SDM has lagged behind. While commercial kits are available for making small point mutations, large deletions/insertions require complicated, often costly in vitro assembly methods. A new method, REPLACR-mutagenesis, harnesses the power of bacterial recombineering to create insertions, deletions, and substitutions - at the same efficiency as Gibson Assembly and GeneArt cloning - but at a much lower cost. Read on to find out how to replace your SDM method with REPLACR (Recombineering of Ends of Linearized Plasmids After PCR).

Current site directed mutagenesis methods
Current SDM methods rely on in vitro assembly. To make a point mutation, you design two oligonucleotide primers with the desired mutation, then use those to PCR amplify the template plasmid. The parental, nonmutated DNA is digested with methylated DNA-specific enzyme DpnI, and the nonmethylated PCR products are then transformed into E. coli to undergo nick repair. This method can work well, but poor template digestion can result in a high number of background colonies carrying the unmodified plasmid. For insertions, it’s possible to use the same technique just using a large primer known as a megaprimer.
Piecing together multiple PCR products using sequence homology is also common. Overlap extension PCR is used to splice together two PCR amplicons based on sequence homology, and the subsequent product is then digested and ligated into an appropriate vector. Gibson Assembly, GeneArt Cloning, and SLIC can all be used to assemble multiple fragments, but these methods can be expensive because they require particular enzymes, some of which are proprietary. These methods also rely on ssDNA ends to match up regions of homology, so they won’t work if the fragment ends have secondary structure or too much sequence similarity.
REPLACR-mutagenesis: Letting bacteria do the work
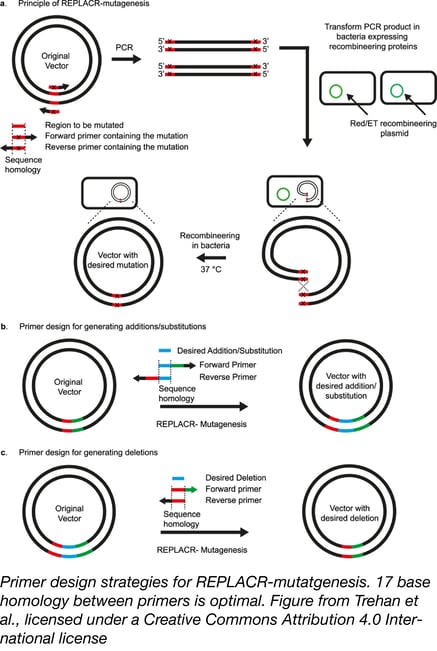 If you’ve conducted SDM using some of these methods, chances are you’d like to find a better one. Trehan et al. wondered if, instead of relying on in vitro assembly, they could make the E. coli do the work by using the principles of recombineering. In recombineering, bacteria express phage recombination machinery, which facilitates easy genetic modification by homologous recombination. In most recombineering strategies, only 3-4 phage proteins are needed and these can be provided via a separate plasmid or be inserted into the bacterial genome.
If you’ve conducted SDM using some of these methods, chances are you’d like to find a better one. Trehan et al. wondered if, instead of relying on in vitro assembly, they could make the E. coli do the work by using the principles of recombineering. In recombineering, bacteria express phage recombination machinery, which facilitates easy genetic modification by homologous recombination. In most recombineering strategies, only 3-4 phage proteins are needed and these can be provided via a separate plasmid or be inserted into the bacterial genome.
REPLACR-mutagenesis starts out in familiar territory, but quickly diverges from previously discussed methods. As described above, you design two primers, each containing the desired mutations, to amplify both strands of a plasmid template. In addition to containing the mutations, the primers also have some homology to each other; ~17 bases is optimal. After being treated with DpnI to digest away the template plasmid, these PCR products are directly transformed into bacteria expressing recombineering proteins Red/ET from a temperature-sensitive plasmid. Homologous recombination then joins the linear PCR products into stable circular plasmids - no nick repair is needed. Since the bacteria are subsequently grown at 37 °C, the recombineering plasmid is not maintained. Recombination efficiency is high enough to negate the need for selection usually required in recombineering.
REPLACR-mutagenesis is also well suited for insertions and deletions. The figure above shows guidelines for primer design; a desired insertion should be present in both primers. When deleting a region, both primers should cover the newly joined ends surrounding the deletion. REPLACR-mutagenesis successfully incorporated a 60-nucleotide insertion; beyond this insertion size, primer synthesis is more expensive than gBlock synthesis, and multifragment assembly methods are preferred. No such restriction applies to deletions; using REPLACR-mutagenesis, Trehan et al. deleted a whopping 144 kb from a bacterial artificial chromosome in just one reaction. Using other methods, such “BAC shaving” would require multiple steps.
One potential pitfall of REPLACR-mutagenesis is that you must PCR amplify the whole plasmid, which could introduce mutations. Using commercially available high-fidelity polymerases such as KOD and Phusion, which are recommended for plasmids up to 25 kb, most cloners should be fine, especially using plasmids in the 10-12 kb range. REPLACR-mutagenesis also requires making your own electrocompetent cells in which the phage recombination machinery is induced using L-arabinose. These cells can be made and frozen in large batches suitable for many experiments.
When REPLACR-mutagenesis goes head-to-head with Gibson assembly and GeneArt cloning, the results are favorable. With 17-bp homology, the observed efficiency of mutagenesis (mutation-containing colonies/total colonies) is about 84% for each of the three methods. However, REPLACR-mutagenesis is much cheaper and faster since you skip the steps of gel purification and in vitro recombination. Across all modifications described in Trehan et al., REPLACR-mutagenesis’ median efficiency is 75%. If these numbers hold up across labs, REPLACR-mutagenesis could save busy scientists a lot of time. Let us know how REPLACR-mutagenesis works in your hands!
| REPLACR-Mutagenesis | Megaprimer PCR | Gibson Assembly | GeneArt Cloning | |
| Types of Mutations | Insertions, deletions, point mutations | Insertions, deletions, point mutations | Insertions, deletions, point mutations | Insertions, deletions, point mutations |
| PCR Reactions | 1 | 1 | 1 per fragment | 1 per fragment |
| In vitro Assembly | No | No | Yes | Yes |
| DpnI Digestion | Yes | Yes | No | No |
| Purified Enzymes Required | No | No | Yes | Yes |
| Lab Made Competent Cells Required | Yes | No | No | No |
Comparison of REPLACR-mutagenesis with other common methods for site directed mutagenesis
References
1. Trehan, Ashutosh, et al. (2016). “REPLACR-mutagenesis: a one-step method for site-directed mutagenesis by recombineering.” Sci Rep 6:19121. PubMed PMID: 26750263. PubMed Central PMCID: PMC4707547.
Additional Resources on the Addgene Blog
- Read about Other Plasmid Cloning Techniques like SLIC and Golden Gate Cloning
- Learn How to Verify Your Plasmid
- Read our Plasmids 101 Series
Additional Resources on Addgene.org
Topics: Other Plasmid Tools, Plasmids






Leave a Comment