Over the past decade, scientists have developed and fine tuned many different ways to clone DNA fragments which have provided appealing alternatives to restriction enzyme cloning. These newer technologies have become more and more common, and for good reason. They offer many advantages over the traditional restriction enzyme cloning we once relied exclusively on. In this blog post, I will go over some advantages, disadvantages, and examples of how scientists are using Gibson assembly to put together DNA fragments.
As a fun way to start, I highly recommend watching this entertaining video created by our friends on the Cambridge 2010 iGEM team that describes the basics of Gibson assembly as a parody of “Breakfast at Tiffany’s.”
Overview of Gibson assembly
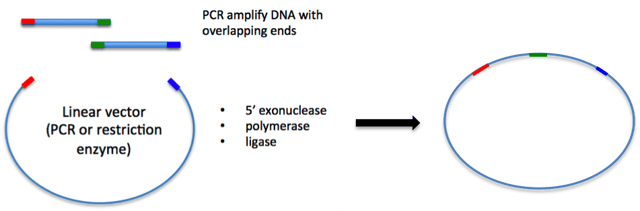
The Gibson assembly technique was first described by Dr. Daniel Gibson and colleagues at the J. Craig Venter Institute in 2009. Here at Addgene we added this option to our drop down menu of common cloning options in the deposit process in 2014 because of its gain in popularity. Gibson assembly is well known for allowing easy assembly of multiple linear DNA fragments, but can also be used in basic cloning of an insert into your vector of choice as shown in the figure below. To start, you need to have DNA fragments with regions of homology at their ends, which are typically created by PCR. Then, the fragments are incubated together with an enzyme master mix, which contains three different enzymes:
- an exonuclease, which chews back the 5’ ends of the fragment, generating long overhangs which allows the single stranded regions with homology to anneal
- a polymerase, to fill in the gaps
- a DNA ligase, to seal the nicks of the annealed and filled-in gaps
The great part about this mix of enzymes is that they can all work at the same temperature, so the entire reaction takes an hour or less to complete at 50 °C. After an hour or so, the sample is immediately ready to transform into competent cells. The master mix of enzymes can be purchased from a company (e.g. NEB or SGI-DNA), or can be mixed yourself (e.g. see the Miller Lab Protocol). The Gibson assembly process can be used to assemble up to 6 fragments in one step, resulting in scar-free assembly that does not require the presence of specific restriction sites (or lack thereof) nor a serious time commitment. Another advantage is that this process makes it easy to generate wild type and mutant constructs at the same time, rather than sequentially.
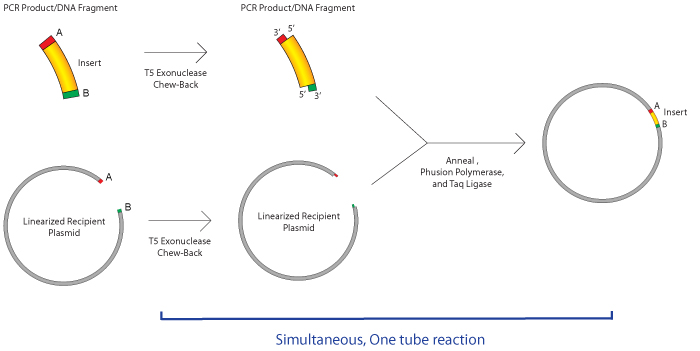
How does Gibson assembly work?
The required homology between neighboring fragments can be created via PCR amplification with primers that contain the appropriate homologous sequences. NEB recommends an overlap of 15-40 bp, with a primer melting temperature greater than 48℃. Both Snapgene and NEB have tools that help you design primers for PCR amplification of fragments to incorporate such regions of homology. This video gives a helpful demonstration of how to use Snapgene’s program to design primers for Gibson Assembly.
For a simple example of using Gibson assembly, imagine that you want to insert your gene of interest into a vector with a large tag at the N-terminus, but you don’t have the tag already included in the vector you want to use. If you inserted these two pieces of DNA by restriction enzyme cloning, you would have to do it in two steps, and a scar would likely remain between the two fragments. However, by using Gibson assembly, you can insert both the gene of interest and the tag sequences into the vector in one step without scars as depicted below. First, you need to design primers to amplify the two fragments while also including regions of homology to the vector or neighboring fragment. Then you would amplify the fragments and vector by PCR, verify that you have bands of the correct size, and purify the DNA fragments. Finally, you simply incubate these three fragments together along with the Gibson assembly master mix for 1 hour, and then transform into competent cells. The success rate of this reaction is typically fairly high, so there is usually no need to screen a large amount of colonies. Excluding the time it takes to get primers, you can have your construct made in 5 days.
One drawback to the Gibson assembly technique is that the process works best with fragments over 200 nucleotides. This may be because the exonuclease could chew through an entire fragment shorter than 200 nucleotides before the annealing and polymerization steps could occur. Secondly, it does not work well if the ends of the fragments have stable single stranded DNA secondary structure, such as a hairpin or a stem loop (as might be anticipated to occur within a terminator sequence), since this would directly compete with the required single-stranded annealing and priming of neighboring assembly fragments.
Gibson assembly is commonly used in synthetic biology, mainly because of the ease in assembling multiple fragments in one step with no scar sequences remaining in the final product. The long overlapping region between fragments also better ensures correct assembly order of the fragments as compared to the smaller overlapping sequences left with a restriction digest. A study in 2013 found that Gibson assembly was one of the most commonly used assembly methods (Kahl 2013). However, Gibson assembly isn’t ideal for synthetic biology standards, which rely up on the re-use of parts between experiments. In Gibson assembly, long primers for each fragment must be designed and ordered, and are specific for each fragment as well the fragment you want next to it, so this doesn’t allow for mix and match of many different fragments. One way around this is to use a combination of standard sequences with overlap regions, such as in the case of MODAL (modular overlap-directed assembly with linkers) which brings modularity to overlap-directed methods (Casini 2014). In this case linker sequences are added before the homology regions, which allows for the mixing and matching of parts.
Gibson assembly meets CRISPR
Gibson can be adapted to more complicated cloning schemes, such as those where the vector that you want to use is very large, has a high GC content, contains a lot of repeats--any of which could make the PCR step difficult--OR there is no convenient restriction site for linearization. This is a perfect case for the use of Gibson assembly combined with the popular CRISPR technology and is described in a recent publication by the Lockey lab (Wang, et al. 2015). In this case, rather than using a restriction enzyme or PCR to make linearized vector, the Cas9 enzyme along with a specific gRNA was used to cleave the 22kb vector. When followed by the standard Gibson assembly techniques described above, this resulted in the direct and seamless cloning into a vector where no other methods were available. A second example of using Gibson Assembly along with CRISPR was also described recently (Jiang et al. 2015), in which very large fragments of the bacterial chromosome (up to 100 kb) were specifically cut out via CRISPR and then assembled into a vector using Gibson assembly. In this case the vector was PCR amplified to contain regions of homology with the bacterial chromosome fragments. CRISPR cutting was used to circumvent the need to PCR amplify the fragments of the chromosome, which was technically challenging.
Other homology based technologies
We've described Sequence and Ligation Independent Cloning (SLIC) in a previous Plasmids 101 post. Although SLIC may be more cost effective, Gibson assembly improves on two aspects of the SLIC methods. First, it uses a dedicated 5’ exonuclease instead of using the exonuclease feature of T4 DNA polymerase, which must be controlled by the presence or absence of dNTPs. Secondly, in Gibson assembly a ligase is added to repair the nicks in vitro, whereas in SLIC these constructs are repaired in vivo, which ends up being much less efficient.
In addition to SLIC and Gibson, there are yet more homology based assembly methods that have been described--CPEC (circular polymerase extension cloning) and SLiCE (Seamless Ligation Cloning Extract) to name two more. Likewise, there are several store-bought cloning kits available which are all based on long-overlapping regions, have no requirements for restriction enzymes, and no scar sequences between fragments. Some of these products include:
- GeneArt® Seamless Cloning and Assembly (Thermo Fisher Scientific)
- NEBuilder® HiFi DNA Assembly (NEB)
- Cold Fusion Cloning (System Biosciences)
- In-fusion Cloning (Clontech)
Although these kits may come at a high price, their manufacturers tout great efficiency and low time commitments. These kits also come with specific protocols, suggestions for ratios of product to insert, and tools for primer design, so it’s always best to check in with the instructions from the particular manufacturer of the kit you will be using. Some of these products may offer advantages that Gibson assembly does not such as increased efficiency, shorter incubation times, or the ability to accommodate smaller fragments.
Know your cloning methods
There are two other common cloning methods that are easy to get mixed up with Gibson assembly (they all start with a G!) but actually work by significantly different methods. Golden Gate cloning does result in the seamless joining of fragments, but uses site specific restriction sites (Type IIS restriction endonucleases) to cleave DNA outside of the recognition sequence. This requires that the vectors and DNA fragments contain these sites at the correct location and NOT in the middle of your insert. Gateway cloning utilizes λ integrase to catalyze directional cloning of DNA parts that are flanked by orthogonal versions of the attB and attP sites recognized by the integrase. This method requires specialized vectors containing these integration site and leaves scars between fragments, but allows for easy movement of a DNA fragment from one vector to another.
There are lots of different ways to clone these days. Gibson and the other long-homology based cloning methods are useful alternatives to the standard restriction/ligation, Gateway, or Golden Gate cloning methods. Whether it is for routine cloning, assembly of multiple fragments, or synthetic biology, you should consider giving it a try!
References:
1. Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009; 6, 343-345. PubMed PMID: 19363495.
2. Wang JW, Wang A, Li K, Wang B, Jin S, Reiser M, Lockey RF. CRISPR/Cas9 nuclease cleavage combined with Gibson assembly for seamless cloning. BioTechniques 2015; 58:161-170. PubMed PMID: 25861928.
3. Jiang W, Zhao X, Gabrieli T, Lou C, Ebenstein Y, Zhu TF. Cas9-Assisted Targeting of CHromosome segments CATCH enables one-step targeted cloning of large gene clusters. Nat Commun. 2015; Sep 1;6:8101. PubMed PMID: 26323354. PubMed Central PMCID: PMC4569707.
4. Kahl, L. J. & Endy, D. A survey of enabling technologies in synthetic biology. J. Biol. Eng. 2013; 7, 13. PubMed PMID: 23663447. PubMed Central PMCID: PMC3684516.
5. Casini, Arturo, et al. "One-pot DNA construction for synthetic biology: the Modular Overlap-Directed Assembly with Linkers (MODAL) strategy." Nucleic acids research 42.1 (2014): e7-e7. PubMed PMID: 24153110. PubMed Central PMCID: PMC3874208
Other Resources:
Additional Resources on the Addgene Blog
- Browse All of Our Plasmids 101 Posts
- Read About Other Cloning Techniques
Additional Resources on Addgene.org
Gibson Assembly® is licensed to New England Biolabs by Synthetic Genomics, Inc.
Topics: Plasmids 101, Plasmid Cloning, Plasmids








Leave a Comment