Around 20% of human genes have no known function or a poorly defined function (Wood, et al). In the microbial world, approximately 50% of predicted genes have unidentified roles as well (Vanni, et al). Is it true that this many genes are truly dispensable or are the current identification methods missing something? Here we will review some time-tested gene identification systems that may have a leg up on their modern counterparts.
What are traps?
Traps – gene, enhancer, and promoter – all rely on the random integration of a reporter construct in close proximity to an endogenous gene. Each trap uses a slightly different approach (described in detail below), but fundamentally the reporter construct monitors the expression of a nearby gene. The reporter gene serves as a proxy for the endogenous gene – its expression levels (high/low) and patterns (tissue/timing) can be monitored under various conditions without necessarily modifying the gene of interest in any way. For example, if green fluorescent protein (GFP) was the reporter gene and green cells were observed in the kidneys at a specific developmental stage you could infer that the gene which GFP was monitoring was being expressed in the same pattern. These systems suggest the type of phenotype to look for with an uncharacterized gene.
The actual reporter construct can be something phenotypic (such as a fluorescent protein or something species-specific) or a selectable marker. Each insertion line can then be isolated and screened. Depending on the marker, traps can be used as high-throughput screens to introduce reporters throughout an organism’s genome. These reporters have historically been used extensively in fly, mouse, and yeast models.
Why traps are useful
Traditional genetic screens employ various on/off switches to measure the function of a gene. For example, they eliminate the expression of the gene (by CRISPR knockout, shRNA depletion, etc.) and ask what the phenotype is without that gene product. These are effective tools but neglect some biologically relevant situations.
- Tissue-specific genes – the genes of interest may not be expressed in the tissue/cell line you are working with.
- Development-specific genes – if a gene functions at multiple stages in development, its depletion may result in lethality before all of its functions can be observed.
- Functionally redundant genes – multiple genes which function in overlapping roles can mask the depletion of one another.
Under these circumstances, trap methods may be the preferred screening option.
Promoter traps
Promoter trap systems lack a promoter and thus require the trapping of an endogenous promoter. The system is composed of a promoter-less reporter which needs to integrate within an exon of a gene to generate a transcriptional fusion. This may disrupt the endogenous function of the gene as a result but will still allow for expression pattern monitoring.
Enhancer traps
Like promoter traps, enhancer traps lack enhancer elements and require the trapping of endogenous enhancers to stimulate the expression of the reporter. The system is made up of a reporter gene flanked by a minimal promoter which can integrate proximal to an endogenous gene to turn on expression. Enhancer traps do not need to be integrated within an intron or exon of a gene, and thus they do not disrupt the gene expression of the gene they monitor.
Gene traps
Gene traps are promotor-less constructs that contain splice acceptors. They require insertion into an intron of a gene, where they can be spliced into the gene product resulting in a transcriptional fusion, much like promoter traps. While this system does not require insertion into an exon, like promoter traps, it will still likely disrupt the gene it monitors due to transcriptional fusion.
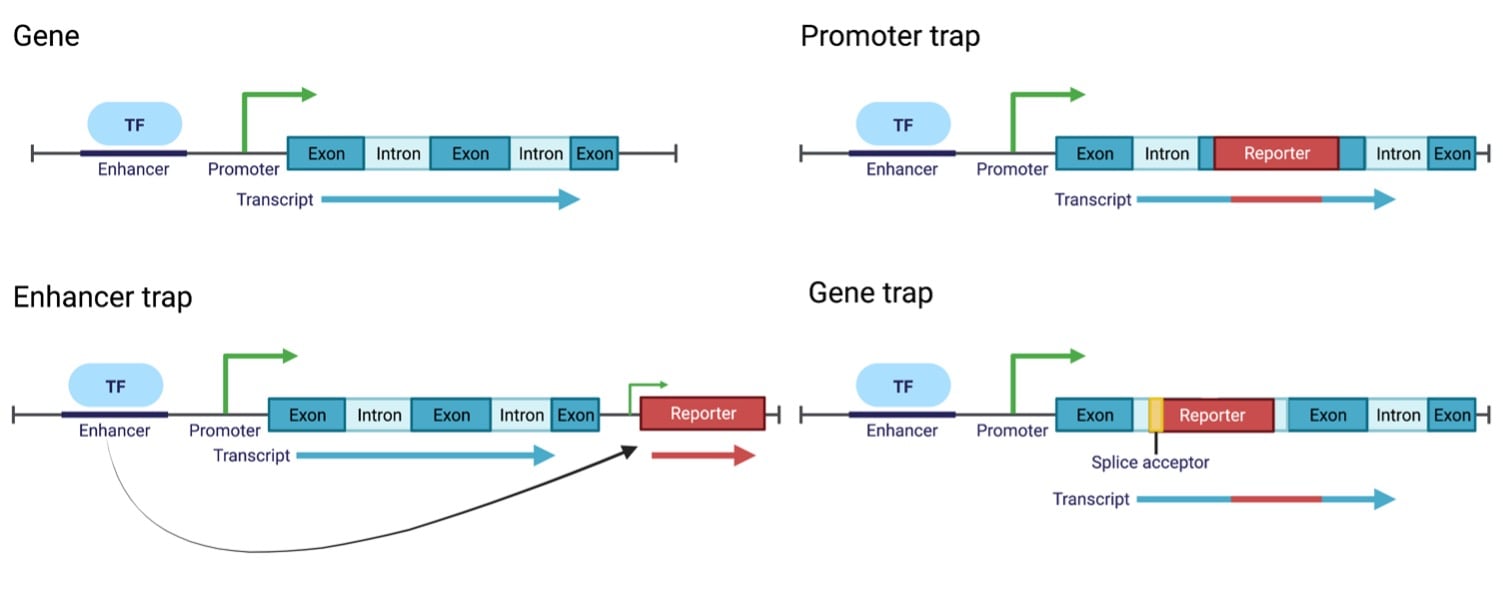
Examples of promoter, enhancer, and gene traps at a genomic locus. |
Choosing a trap
At first glance, you may think enhancer traps are the top trap – they don’t require insertion in a highly specific gene region and they don’t disrupt the expression of the endogenous gene. These are the pros of enhancer traps! However, don’t count the other traps out just yet. Gene and promoter traps unambiguously identify the gene that they are monitoring as they are directly integrated into it. Enhancer traps do not need to be inside of a gene’s open reading frame, in fact, they can be activated a considerable distance away from a gene which can make identification of the gene itself difficult. The best trap depends on your experiment and the organism you are working in. The spacing of genes and whether or not it’s ok to disrupt endogenous gene expression are all factors to consider.
Once you’ve decided on the type of trap, there are many ways to insert your construct to generate a collection of monitored genes and mutants - viral integration, transposon, and many more! Check out Addgene’s catalog to find trapping tools for your next experiment!
References and Resources
References
Wood, V., et al., Hidden in plain sight: what remains to be discovered in the eukaryotic proteome? Open Biol. (2019) 9:180241. DOI: https://doi.org/10.1080/rsob.180241
Vanni, C., et al., Unifying the known and unknown microbial coding sequence space. eLife. (2022) 11:e67667. DOI: https://doi.org/10.7554/eLife.67667
Springer, S. P. Gene Traps. Plant Cell. (2000) 12(7): 1007-1020. DOI: 10.1105/tpc.12.7.1007
Additional resources on Addgene.org
Additional resources on the Addgene blog
Topics: Plasmids 101





Leave a Comment