You’ve committed to the daunting task of finding new factors in your biological system of interest, the so-called “fishing expedition.” Your first question is: to screen or to select? Which method will work best for your scientific question? If you’re thinking, ‘wait, those terms mean different things?!’, you aren’t alone! We’ve noticed a lot of scientists struggling with when to use a screen vs. a selection and we are here to discuss the ins and outs of both approaches.
Screens and selections
The common goals
The confusion in differentiating between a screen and a selection isn’t surprising - they have a lot in common. In fact, selections are often inappropriately categorized as screens! Why the confusion? The commonality between these methods is that both attempt to identify new players in a biological process. Players can be proteins, genes, enzymes, drugs, or anything else that plays a role or affects a function in a given process. The process can range from a defined pathway or reaction to a complex event such as cancer or even aging. For both a screen and a selection, it is important to clearly define what type of factor you are looking for and what process or outcome that factor functions in.
The differences
They say the devil is in the details - and that’s where the differences in these approaches can be found. As an example, say you are considering either a cellular screen or a selection. A selection is a process in which only cells with the trait of interest will survive in the selected growth conditions. For screens, there is no obligatory growth advantage – that is, you are not specifically looking at cells that can survive. Instead, all manipulations are assessed, regardless of whether they enhance, hinder, or have no effect on the process you are interested in.
What does this actually look like? For a selection, you might put cells into a drug-containing media in which only cells which can tolerate the drug survive and everything else dies off. For a screen, you would individually treat cells with many different drugs and then perform an assay to look at an outcome. Your scientific question can help determine which is right for you.
Screen variations
Forward and reverse screens
Forward and reverse screens involve manipulating or mutagenizing a population (typically cells or small organisms) and then assaying for a functional or phenotypical outcome. In forward screens, there is random or unknown manipulation of the population. This is contrary to reverse screens, where there is removal of known factors (such as a genes) and the loss of those factors is assessed. The two most common methods of manipulation are genetic knockout mutations and treatment with small molecules/drugs.
With the advent of CRISPR, genome-wide knockout screens are possible with gRNA libraries targeting all known genes. This is a perfect example of an unbiased, reverse screen – all known genes are assayed for their role in a particular capacity. Drug screens, on the other hand, are often performed as biased forward screens. A given panel of drugs are individually administered and then outcomes are measured. While the entire genome can be knocked out with CRISPR, the entirety of the small molecule realm is typically not feasible to test in any given screening experiment. Thus, a limited panel is usually selected.
The method you choose to assess traits of interest is highly specific to your scientific question. Things to consider when selecting an assay system include: How specific is it? Is it high throughput? Will it be sensitive enough? No one wants to spend time screening hits that aren’t “real” or novel for their purposes… or worse yet have a screen that doesn’t yield any hits. Make sure you plan ahead and consider these questions to save yourself a headache later on.
Enhancer and suppressor screens
These types of screens are very similar to forward and reverse screens, except that they begin with a uniform population possessing a specific mutation or phenotype. For example, a drug screen may be performed on a cell line possessing a patient mutation to assess how patients may respond to drug treatment. Another example is using a mouse model of a particular disease as the input population. For these experiments, the goal is to identify manipulations which either enhance or suppress the previously defined disease phenotype. These screens employ the same downstream manipulations as a forward or reverse screen (CRISPR, drug, etc.), just with an added baseline component.
Alternative screening techniques
While CRISPR and drugs dominate the screening scene, there are other options which are worth considering. RNAi screens are similar to CRISPR, but instead of knocking out the genes of interest (or the whole genome), they knock down gene expression temporarily. RNAi libraries are especially useful if you are interested in studying genes whose knockout would be lethal to cells and thus not assayable with CRISPR. Another method, popularized in yeast, are temperature sensitive screens. These involve raising or lowering the culture temperature to exacerbate a mutant phenotype, very similar to enhancer/suppressor screens.
Selections
Selections differ from screens in that only survivable manipulations are assessed, rather than all manipulation events. To put it another way, only cells or organisms with the desired phenotype will survive and be assayed at the end of the selection process. Non-functional traits will be automatically eliminated, unlike in screens. Selections begin and end with the same steps as screens – they start with manipulations (CRISPR, etc.), and finish with the characterization of a function or phenotype in the surviving population. Selections do require there to be a culture condition that can apply enough pressure to select for variants of interest, though. A selection can take the form of varied temperature, nutrient availability, drugs, and much more.
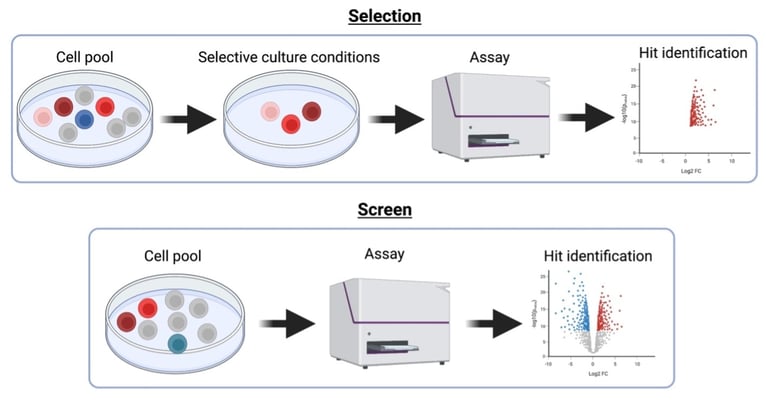 |
| Workflow of screens and selections |
Choosing between selections and screens
Selections sound like a great shortcut, right? Who would ever do a screen when a selection can get you to the answer that much faster? In reality, many biological questions are not conducive to selections and have to be addressed with screens. Selections require there to be a culture condition which eliminates manipulations that do not affect the function of interest. An example where a selection would work is if you are trying to identify genetic conditions which convey resistance to a drug. You could culture cells in the drug of interest at a lethal dose for non-manipulated cells and then assay cells which survive as these must have acquired a favorable mutation. But if the cellular pathway of interest conveys no significant survival advantage, such as a non-essential mechanism or any pathway with cellular redundancies, a screen would be required to answer your experimental question. Now that you understand the difference between the two, it’s time to check out our CRISPR library resources or our RNAi resources and go fishing!
References and resources
References
Wang, T., Wei, J.J., Sabatini, D.M., Lander, E.S. Genetic screens in human cells using the CRISPR/Cas9 system. Science, 343(6166): 80-84 (2014). doi: https://doi.org/10.1126/science.1246981.
Leemhuis, H., Kelly, R.M., Dijkhuizen, L. Directed evolution of enzymes: Library screening strategies. IUBMB Life, 61: 222-228 (2009). doi: https://doi.org/10.1002/iub.165.
Additional resources on the Addgene Blog
Topics: Plasmids 101





Leave a Comment