When performing restriction digests on plasmids at Addgene, we sometimes observe something odd in our uncut DNA control: a band or two appear on an agarose gel at notably higher molecular weights than expected, given the size of the plasmid. This is seen only in the uncut DNA; the rest of the digest appears normal and produces the expected fragments for that plasmid. This phenomenon indicates the presence of a multimer.
 |
|
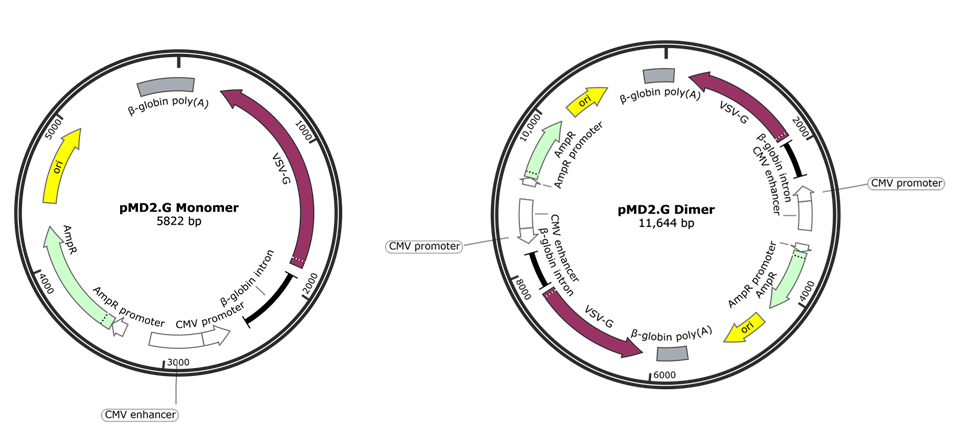
Fig. 1: The monomer of Plasmid 12259: pMD2.G (depicted on the left) is ~5.8 kb in length. The plasmid can exist on as a dimer, depicted on the right, with the entire monomeric sequence duplicated in tandem, resulting in a plasmid that is twice the length of the monomer (~11.6 kb). Image generated by SnapGene software. |
Thanks to increasing affordability and accessibility, more researchers have been utilizing full plasmid sequencing services to verify the plasmids, including long-read sequencing techniques, such as Oxford Nanopore sequencing or PacBio SMRT sequencing, (which differ from the short-read Illumina MiSeq sequencing that Addgene uses.) However, when some researchers received the sequencing results for their plasmids, they found the plasmids were much longer than expected, and the sequence was repeated two or more times in tandem. Such plasmids are known as multimers, and they are the result of multiple plasmid copies combining in a process known as plasmid multimerization.
Plasmid Multimerization
Early studies on plasmid topology recorded plasmid multimerization in the naturally occurring E. coli plasmid ColE1 (Goebel & Helinski, 1968; Bazaral & Helinski, 1968), in which the plasmid was observed to exist in various multimeric forms (e.g. concatemers, dimers, trimers, tetramers). Since then, multimerization has been observed to occur in many different kinds of plasmids, taking many forms (Levene, 2009; Higgins & Vologodskii, 2016). Plasmid multimerization occurs via homologous recombination between two or more copies of the same plasmid (Bedbrook & Ausubel, 1976). This recombination event produces a multimeric form of the plasmid with repeating sequences. In some specific cases, multimerization has been observed to occur more frequently in plasmids with large inserts (>7 kb)(Berg et al., 1989), direct repeats (Ribeiro et al., 2009), and at high copy numbers (Williams, Carnes, and Hodgson, 2009). More research is ongoing to determine plasmid features that affect the frequency of multimerization.
Theoretically, as long as all the plasmid features remain intact, plasmid multimers function just as well as monomers. However, due to their larger size, multimers may exhibit reduced transformation and transfection efficiency compared to the monomeric version of the plasmid (Maucksch et al., 2009). Since multimers have additional origins of replication compared to monomers, they are also able to replicate at a higher frequency (Summers & Sherratt, 1984; Summers et al., 1993). These larger plasmids are also maintained at lower copy numbers within bacterial cells, and bacterial cells with multimers grow more slowly compared to those containing the monomeric version of the plasmid (Summers & Sherratt, 1984; Summers et al., 1993; Summers, 1998). Multimeric plasmids are also considered to be less stable and more prone to being lost in bacterial cells compared to monomers, although this can be prevented via antibiotic selection (Crozat, 2014).
How to Detect Plasmid Multimers
Run an undigested plasmid on agarose gel
Running undigested DNA on a gel as a control when performing a diagnostic digest is a simple but effective method for detecting plasmid multimers. Digests themselves cannot be used to detect multimers, since restriction sites in multimers usually repeat in tandem along with the repeated plasmid sequence, giving greater concentration of cut DNA without changing the size. As a result, a restriction digest of multimers will produce the same fragment sizes and banding patterns on an agarose gel as the monomeric version. However, undigested multimers are larger than monomers and will therefore run higher on an agarose gel (Bedbrook & Ausubel, 1976). In fact, if your plasmid sample has a mix of different-sized multimers, you may even see different sets of bands on the gel corresponding to each type of multimer present in your sample. In these instances, please note that the supercoiled dimer band can sometimes overlap with the band corresponding to the nicked or relaxed conformation of the plasmid monomer (see this article from BiteSize Bio for more information on how different plasmid conformations run on an agarose gel). If the presence of a multimer in your sample remains unclear after visualizing it on a gel, one of the other methods below should help in multimer identification.
|
|
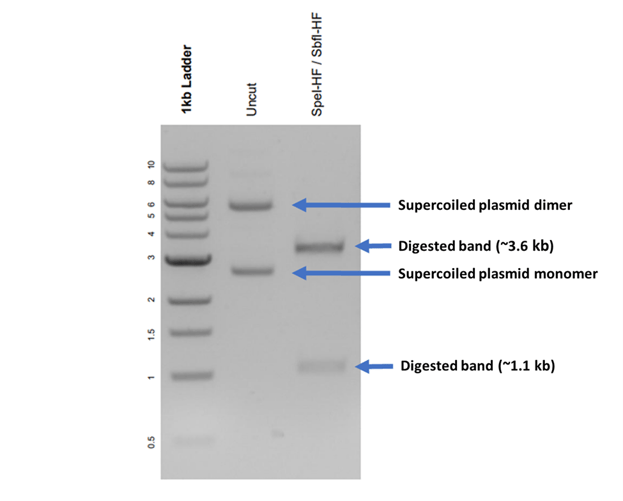
Fig. 2: Gel image of Plasmid 61564: pBAMD1-2 (~4.7 kb) digested with SpeI and SbfI with expected bands at ~3.6 kb and ~1.1 kb (lane 3). Uncut DNA (lane 2) has a band at <3 kb, which corresponds to the supercoiled plasmid monomer, and at <6 kb, consistent with the size of a supercoiled plasmid dimer. Please note that it is common to see other plasmid confirmations like linear and nicked plasmids, as described in this article. |
Long-read sequencing
Addgene uses Illumina MiSeq NGS to perform whole plasmid sequencing of our plasmid samples during our quality control process. Since MiSeq involves sequence assembly from short reads (<300 bp) which cannot distinguish between large repeat sequences within a plasmid, it cannot be used to detect plasmid multimers. Some companies provide NGS whole plasmid sequencing services using long-read sequencing (1-25 kb), such as Oxford Nanopore sequencing or PacBio SMRT sequencing. These longer reads are able to detect large repeat sequences within a plasmid and, therefore, allow for the identification of plasmid multimers.
Capillary Gel Electrophoresis (CGE) analysis
Another technique that can detect multimers is capillary gel electrophoresis (CGE), which has been used in plasmid vaccine and gene therapy development to examine plasmid topologies (e.g. supercoiled, open circular, linearized, etc.) (Schleef et al., 2006; Holovics et al., 2010; Cook et al., 2020). CGE uses laser-induced fluorescence (CGE-LIF) for detection and can provide data on the quantity of various topologies and multimers present within a plasmid sample.
Troubleshooting Tips
|
|
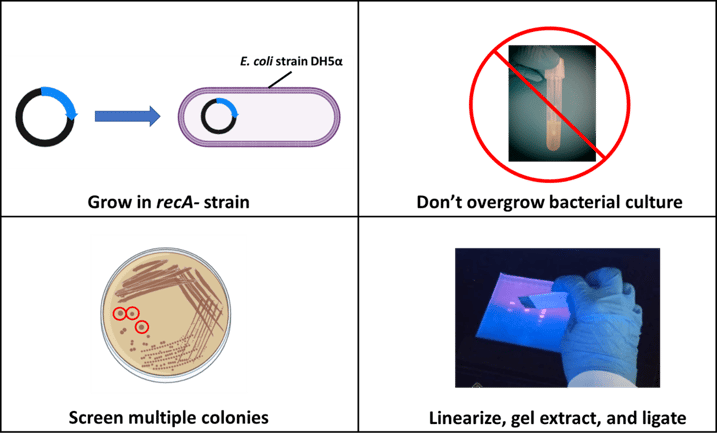
Figure 3. Summary of plasmid multimerization troubleshooting tips. Portions of this figure were made with BioRender. |
Grow plasmids in recombinase-deficient strains
Since plasmid multimerization is caused by homologous recombination, growing a plasmid in a bacterial strain that is deficient in recombinase protein RecA can reduce the frequency of multimerization, and in some cases completely prevent the formation of plasmid multimers (Bedbrook & Ausubel, 1976; Fishel et al., 1981). E. coli strains such as DH5α, HB101, NEB Stable, Stbl3, etc., are recA- mutant strains, and therefore should have reduced frequencies of plasmid multimerization. One study by Bacolla et al. (2011) indicated that strain HB101 produced the highest yield of monomeric plasmid compared to DH5α. However, more research is needed to determine which recA- strains are most effective at reducing multimerization and define recA-independent mechanisms.
Avoid overgrowing the bacterial culture
Another way to prevent plasmid multimerization is to avoid growing the transformed bacterial culture into the late stationary phase. Growing the culture at a lower temperature such as 30°C or optimizing the incubation time can help maintain cultures in log phase growth. In a study by Williams et al. (2009), pUC multimerization was reduced by growing seed stocks at 30°C compared to 42°C.
Screen multiple colonies to isolate the monomer
Sometimes a plasmid can exist in a bacterial population as a mixture of monomers and various forms of multimers (Summers & Sharratt, 1984; Summers et al., 1993). In these cases, screening multiple bacterial colonies using one of the detection methods described above can help ensure the isolation of the monomeric version of the plasmid.
Linearize, gel extract, and ligate the plasmid
If you are having difficulty isolating a plasmid monomer from a plasmid sample, or if no monomers exist in your sample, you could try linearizing the plasmid via restriction digestion and then run the linearized plasmid on an agarose gel. You can then gel extract the linearized band, ligate the plasmid, and re-transform the ligated plasmid to recover the monomeric form of the plasmid.
Have you encountered any multimers in your plasmid samples? What are your strategies for resolving multimers? Let us know in the comments below or contact us at help@addgene.org.
Resources and References
References
Bacolla A, Wang G, Jain A, Chuzhanova NA, Cer RZ, Collins JR, Cooper DN, Bohr VA, Vasquez KM. Non-B DNA-forming sequences and WRN deficiency independently increase the frequency of base substitution in human cells. J Biol Chem. 2011 Mar 25;286(12):10017-26. doi: 10.1074/jbc.M110.176636. Epub 2011 Feb 1. PMID: 21285356; PMCID: PMC3060453.
Bazaral M, Helinski DR. Characterization of multiple circular DNA forms of colicinogenic factor E-1 from Proteus mirabilis. Biochemistry. 1968 Oct;7(10):3513-20. doi: 10.1021/bi00850a028. PMID: 4878700.
Bedbrook JR, Ausubel FM. Recombination between bacterial plasmids leading to the formation of plasmid multimers. Cell. 1976 Dec;9(4 PT 2):707-16. doi: 10.1016/0092-8674(76)90134-3. PMID: 797459.
Berg CM, Liu L, Coon M, Strausbaugh LD, Gray P, Vartak NB, Brown M, Talbot D, Berg DE. pBR322-derived multicopy plasmids harboring large inserts are often dimers in Escherichia coli K-12. Plasmid. 1989 Mar;21(2):138-41. doi: 10.1016/0147-619x(89)90057-7. PMID: 2544913.
Cook KS, Luo J, Guttman A, Thompson L. Vaccine Plasmid Topology Monitoring by Capillary Gel Electrophoresis. Curr Mol Med. 2020;20(10):798-805. doi: 10.2174/1566524020666200427110452. PMID: 32338220.
Crozat E, Fournes F, Cornet F, Hallet B, Rousseau P. Resolution of Multimeric Forms of Circular Plasmids and Chromosomes. Microbiol Spectr. 2014 Oct;2(5). doi: 10.1128/microbiolspec.PLAS-0025-2014. PMID: 26104344.
Fishel RA, James AA, Kolodner R. recA-independent general genetic recombination of plasmids. Nature. 1981 Nov 12;294(5837):184-6. doi: 10.1038/294184a0. PMID: 7029308.
Goebel W, Helinski DR. Generation of higher multiple circular DNA forms in bacteria. Proc Natl Acad Sci U S A. 1968 Dec;61(4):1406-13. doi: 10.1073/pnas.61.4.1406. PMID: 4884687; PMCID: PMC225270.
Higgins NP, Vologodskii AV. Topological Behavior of Plasmid DNA. Microbiol Spectr. 2015 Apr;3(2):10.1128/microbiolspec.PLAS-0036-2014. doi: 10.1128/microbiolspec.PLAS-0036-2014. PMID: 26104708; PMCID: PMC4480603.
Holovics HJ, He Y, Lacher NA, Ruesch MN. Capillary gel electrophoresis with laser-induced fluorescence of plasmid DNA in untreated capillary. Electrophoresis. 2010 Jul;31(14):2436-41. doi: 10.1002/elps.201000061. PMID: 20589859.
Levene SD. Analysis of DNA topoisomers, knots, and catenanes by agarose gel electrophoresis. Methods Mol Biol. 2009;582:11-25. doi: 10.1007/978-1-60761-340-4_2. PMID: 19763938.
Maucksch C, Bohla A, Hoffmann F, Schleef M, Aneja MK, Elfinger M, Hartl D, Rudolph C. Transgene expression of transfected supercoiled plasmid DNA concatemers in mammalian cells. J Gene Med. 2009 May;11(5):444-53. doi: 10.1002/jgm.1310. PMID: 19263463.
Ribeiro SC, Oliveira PH, Prazeres DM, Monteiro GA. High frequency plasmid recombination mediated by 28 bp direct repeats. Mol Biotechnol. 2008 Nov;40(3):252-60. doi: 10.1007/s12033-008-9082-3. Epub 2008 Jul 8. PMID: 18607781.
Schleef M, Baier R, Walther W, Michel ML, Schmeer M. Long-term stability study and topology analysis of plasmid DNA by capillary gel electrophoresis. BioProcess Int. 2006 Sep;4(8):38-40.
Summers D. Timing, self-control and a sense of direction are the secrets of multicopy plasmid stability. Mol Microbiol. 1998 Sep;29(5):1137-45. doi: 10.1046/j.1365-2958.1998.01012.x. PMID: 9767582.
Summers DK, Beton CW, Withers HL. Multicopy plasmid instability: the dimer catastrophe hypothesis. Mol Microbiol. 1993 Jun;8(6):1031-8. doi: 10.1111/j.1365-2958.1993.tb01648.x. PMID: 8361350.
Summers DK, Sherratt DJ. Multimerization of high copy number plasmids causes instability: CoIE1 encodes a determinant essential for plasmid monomerization and stability. Cell. 1984 Apr;36(4):1097-103. doi: 10.1016/0092-8674(84)90060-6. PMID: 6323019.
Williams JA, Carnes AE, Hodgson CP. Plasmid DNA vaccine vector design: impact on efficacy, safety and upstream production. Biotechnol Adv. 2009 Jul-Aug;27(4):353-70. doi: 10.1016/j.biotechadv.2009.02.003. Epub 2009 Feb 20. PMID: 19233255; PMCID: PMC2693335.
Williams JA, Luke J, Langtry S, Anderson S, Hodgson CP, Carnes AE. Generic plasmid DNA production platform incorporating low metabolic burden seed-stock and fed-batch fermentation processes. Biotechnol Bioeng. 2009 Aug 15;103(6):1129-43. doi: 10.1002/bit.22347. PMID: 19408315; PMCID: PMC2735187.
More resources on Addgene.org
Addgene's Plasmids 101 eBook
Addgene's Molecular Biology Reference
Addgene's Restriction Digest Protocol
More resources on the Addgene blog:
Plasmids 101: What is a plasmid?
Plasmids 101: Origin of Replication
Plasmids 101: PCR
Topics: Plasmids 101, Plasmids





Leave a Comment