In the last decade, the use of degron tags has become increasingly popular for the modulation of endogenously and exogenously expressed proteins. Here we will review what advantages degron tags can offer over other protein control methods and compare the commonly available types of degron tags.
What is a degron tag and why should I use one?
Degron tags are a class of protein tag which can be added to either the N or C terminal of a protein and can inducibly facilitate the degradation of the tagged protein. Degrons are destabilizing domains which will be identified by various enzymes and sent to the proteasome for degradation. The tags we will discuss here are all conditional, meaning that they only facilitate degradation of their fused protein under certain conditions (typically the addition of an exogenous small molecule). Degron tags can range in size from several kDa up to ~35 kDa and are typically knocked-in via gene targeting or cloned in-frame to the protein of interest for plasmid-based expression systems.
Degron tags are one of many tools to manipulate protein expression. These tags offer several advantages over the alternative systems – CRISPR, siRNA, etc. Firstly, they are reversible mechanisms of control. Protein degradation can be induced by the addition of a small molecule, and protein levels can return to normal once the drug has been removed. The degradation and reversal are rapid compared to other systems because the system modulates protein post-translationally, unlike many other methods. Secondly, these tags can be used to manipulate endogenously and exogenously expressed proteins. Thirdly, degrons and their drugs are titratable. With these tags, protein levels aren’t limited to ‘on’ and ‘off’; varied levels of drug concentration can yield intermediate protein abundance. Finally, some degron tags can do more than just degrade! Read on to find out more.
Common degron tags
When it comes to degrons, you have options! Below we will review the 4 most common types of degrons and some of the nuances to each one.
SMASh
Small molecule-assisted shutoff (SMASh) functions on the basis of both a degron and a viral protease. The 34 kDa tag is oriented such that the viral protease is situated between the degron and the gene of interest and the degron domain is terminal. Between the tag elements (protease and degron) and the protein of interest, there is also a protease recognition site. In unperturbed conditions, the protease cleaves itself and the degron off at the protease cleavage sequence. The tag is then shuttled to the proteasome due to the presence of the degron, and the protein of interest is untagged and functions per the usual. In the presence of a specific protease inhibitor, asunaprevir, the viral protease can no longer self-cleave and the protease and degron remain attached to the tagged protein. In this event, the degron domain shuttles the entire tagged protein to the proteasome for degradation.
In addition to having the coolest name of all the tags reviewed here, the SMASh tag does offer a few other unique features. If your protein is not amenable to being tagged, e.g. tagging some proteins can affect their function, the SMASh tag self-cleaves itself when you want the protein around, leaving a minimal additional sequence (7 kDa). This system also requires less genetic engineering than some of the other systems reviewed here. The only required components are 1) a SMASh-tagged protein and 2) asunaprevir (small molecule).
dTAG
The degradation tag (dTAG) system functions by harnessing the power of a novel degrader of the 12 kDa protein FKBP12F36V. By fusing FKPB12F36V to your protein of interest, it can be degraded by this selective degrader. The novel degrader, dTAG13, is a small molecule which induces dimerization of the FKPB12F36V fusion protein with the endogenous CRBN E3 ligase complex which will polyubiquitinate the fusion protein. This will target the protein of interest to the proteasome for degradation.
Like several other tags described here, the dTAG uses a small molecule, a tag, and the endogenous cellular degradation machinery to function. The dTAG is the smallest tag reviewed here, in regard to total tag size, which can also be handy!
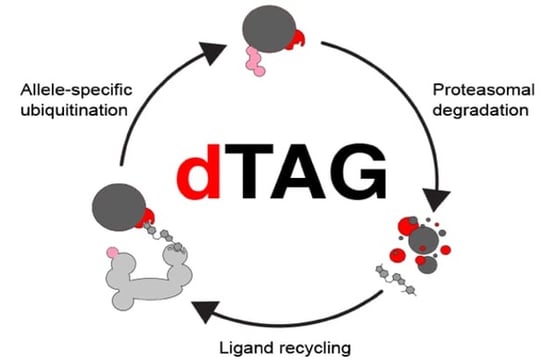
|
|
Fig 1. dTag molecules induce dimerization of an FKBP12F36V fused target protein-of-interest and the CRBN E3 ligase complex leading to ubiquitination and proteasomal degradation. Adapted from Nabet et al., (2018). |
HaloTag
The HaloTag was initially developed by Promega as an affinity tag for visualization of proteins of interest. A fluorescent Halo ligand can be added to media, buffer, etc., which will bind and light up a tagged protein for imaging. Given its popularity and widespread usage, a proteolysis-targeting chimera (PROTAC) was developed for HaloTag. PROTACs are engineered molecules which target E3 ligases to specific proteins by simultaneously binding both their target protein and an E3 ligase through interacting ligands. After an E3 ligase facilitates ubiquitination of the Halo-tagged protein in question, it is targeted to the proteasome for degradation.
The HaloTag is quite large (33 kDa) and does not self-cleave off of the protein like the SMASh tag does. However, it is a straightforward system — HaloTag and its PROTAC are all that is needed to facilitate degradation. HaloTag is also the degron tag with the most alternative functions — there are antibodies to it and ways to achieve fluorescence with it.
AID
The auxin-inducible degron (AID) system is the oldest system reviewed here. The AID system relies on three components. First, the protein of interest must be AID-tagged with the 25 kDa tag. Next, the TIR1 F-box protein must be expressed (inducibly, ectopically, stably… it’s your call). The TIR1 F-box protein will associate with the endogenously expressed SCF complex members, an E3 ubiquitin ligase complex. Finally, when auxin is added in the form of auxin hormone indole-3-acetic acid (IAA), the TIR1 F-box associated E3 ligase complex will ubiquitinate the AID-tagged protein which will target it for proteasomal degradation.
Since its first use as a protein regulation tool, AID has gone through an additional round of evolution (AID-2) to reduce some of the leakiness associated with the system, but the general concept of the system has remained the same. An obvious drawback of the system is that it requires an additional component to be engineered or expressed beyond the tag and small molecule. It does work in mammalian cells, mice, and even yeast, though (we have a kit!).
Choosing a degron tag
Are all tags created equal? No! Choosing the ‘right’ tag does depend a lot on your unique experimental needs. If your protein’s function is easily perturbed by tags, then a small tag might be the way to go (dTAG/SMASh). If you don’t want to deal with additional components beyond the minimum required for a degron system, then it’s probably best to avoid AID tags. If your project also involves imaging at some stage, then choosing a HaloTag could pay dividends down the road.
It is difficult to predict precisely how a given degron tag will cooperate with your protein of interest and how effective it will be, given the diversity of protein structures, interacting partners, and localization. Thankfully, this is a hot topic with many resources! Bondeson et al. (2022) reviews all the tags discussed here and profile their effectiveness with over a dozen different proteins. In the end, you can’t predict with absolute certainty how a degron will behave with a new protein, but you can make an informed choice!
References and Resources
References
Bondeson, D.P., Mullin-Bernstein, Z., Oliver, S. et al. Systematic profiling of conditional degron tag technologies for target validation studies. Nat Commun. 13, 5495 (2022). doi: https://doi.org/10.1038/s41467-022-33246-4
Chung HK, Jacobs CL, Huo Y, Yang J, Krumm SA, Plemper RK, Tsien RY, Lin MZ. Tunable and reversible drug control of protein production via a self-excising degron. Nat Chem Biol. 2015 Sep;11(9):713-20. doi: https://doi.org/10.1038/nchembio.1869
Nabet B, Roberts JM, Buckley DL, Paulk J, Dastjerdi S, et al. The dTAG system for immediate and target-specific protein degradation. Nat Chem Biol. 2018 May;14(5):431-441. doi: https://doi.org/10.1038/s41589-018-0021-8
Holland AJ, Fachinetti D, Han JS, Cleveland DW. Inducible, reversible system for the rapid and complete degradation of proteins in mammalian cells. Proc Natl Acad Sci U S A. 2012 Dec 4;109(49):E3350-7. doi: https://doi.org/10.1073/pnas.1216880109
Resources on Addgene.org
Resources on the Addgene blog
- Plasmids 101: Protein Tags
- Plasmids for Endogenous Gene Tagging in Human Cells
- Antibodies 101: Epitope Tags
Topics: Plasmids 101






Leave a Comment