 In previous posts for our Plasmids 101 series, we examined a number of important plasmid elements – promoters, origins of replication, protein tags, and antibiotic resistance markers (just to name a few). In this edition, we’re going to take a look at a very interesting tool that can be used for creating (excuse the pun) specific, targeted DNA modifications in transgenic animals, embryonic stem cells, and/or tissue-specific cell types: Cre-lox recombination.
In previous posts for our Plasmids 101 series, we examined a number of important plasmid elements – promoters, origins of replication, protein tags, and antibiotic resistance markers (just to name a few). In this edition, we’re going to take a look at a very interesting tool that can be used for creating (excuse the pun) specific, targeted DNA modifications in transgenic animals, embryonic stem cells, and/or tissue-specific cell types: Cre-lox recombination.
What is Cre-lox?
The Cre-lox system is a technology that can be used to induce site-specific recombination events. The system consists of two components derived from the P1 bacteriophage: the Cre recombinase and a loxP recognition site. The P1 bacteriophage uses these components as part of its natural viral life-cycle, and researchers have adapted the components for use in genome manipulation.
Cre recombinase, originally named because it “causes recombination” (although later referred to as the “cyclization recombinase”), is a 38 kDa protein responsible for intra- and inter-molecular recombination at the loxP recognition sites. A key advantage of the system is that Cre acts independently of any other accessory proteins or co-factors, thus allowing for broad applications in a variety of experiments.
LoxP (locus of X(cross)-over in P1) sites are 34-base-pair long recognition sequences consisting of two 13-bp long palindromic repeats separated by an 8-bp long asymmetric core spacer sequence. The asymmetry in the core sequence gives the loxP site directionality, and the canonical loxP sequence is ATAACTTCGTATA-GCATACAT-TATACGAAGTTAT. The loxP sequence does not occur naturally in any known genome other than P1 phage, and is long enough that there is virtually no chance of it occurring randomly. Therefore, inserting loxP sites at deliberate locations in a DNA sequence allows for very specific manipulations as discussed below.
How does it work?
As alluded to above, the Cre recombinase catalyzes the site specific recombination event between two loxP sites, which can be located either on the same or on separate pieces of DNA. Both 13bp repeat sequences on a single loxP site are recognized and bound by a Cre protein, forming a dimer. The two loxP sites then align in a parallel orientation, allowing the four Cre proteins to form a tetramer. A double-strand DNA break occurs within the core spacer of each loxP site and the two strands are ligated, resulting in the reciprocal crossover event.
This event can have three general outcomes based on the location and orientation of the loxP sites:
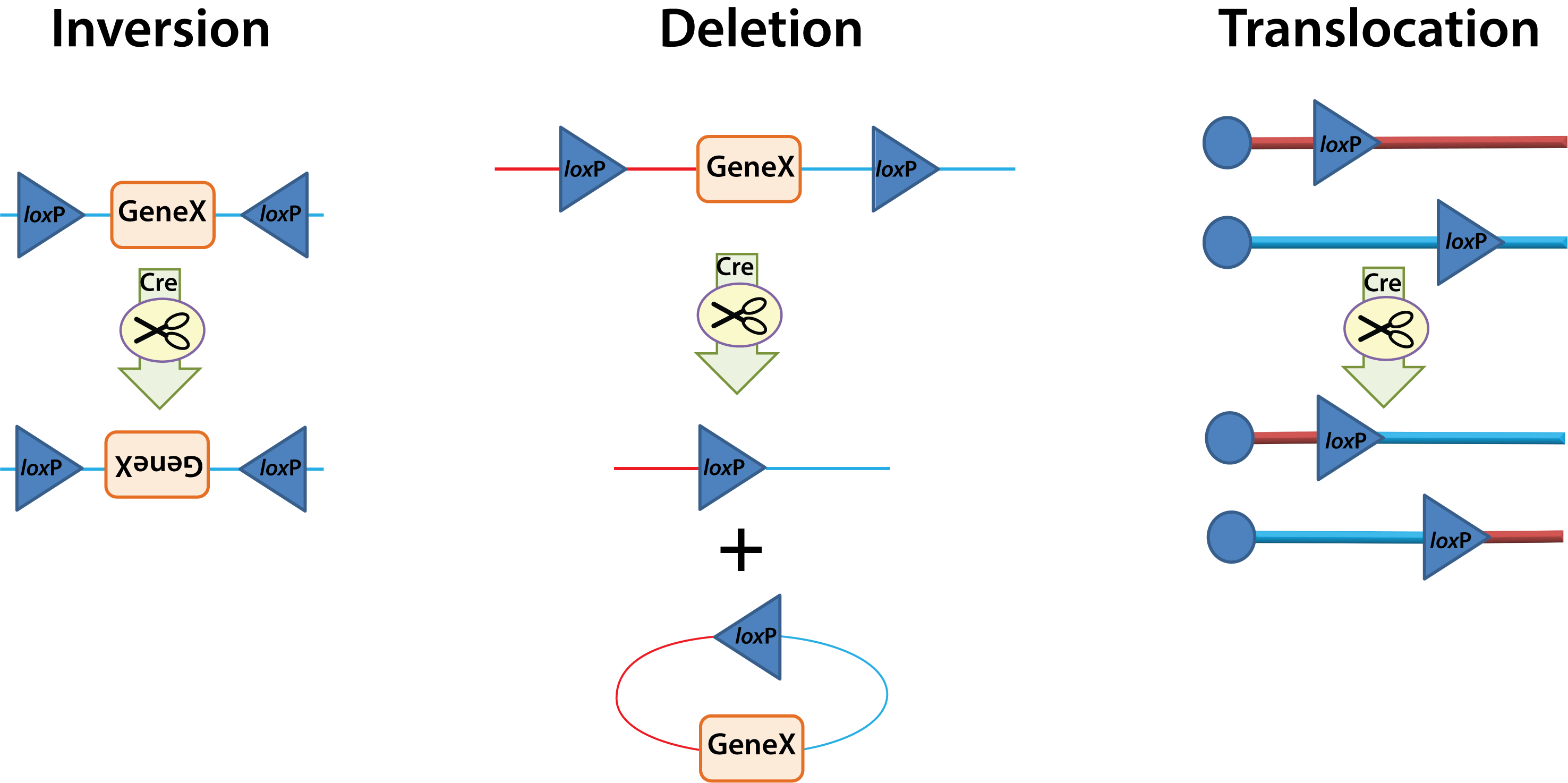
Inversion: If the loxP sites are on the same DNA strand and are in opposite orientations, recombination results in an inversion and the region of DNA between the loxP sites is reversed.
Deletion: If the sites face in the same direction, the sequence between the loxP sites is excised as a circular piece of DNA (and is not maintained).
Translocation: If the sites are on separate DNA molecules, a translocation event is generated at the loxP sites.
How can I use Cre-lox?
The Cre/lox system is a well-established research tool, especially in the field of mouse transgenics. A few of the most common uses are listed below.
- Cre-dependent gene expression – placing a series of transcriptional and translational termination features with loxP sites on either side (often called a “lox-stop-lox” or “LSL” cassette) upstream of a gene of interest will prevent gene expression in the absence of Cre. In the presence of Cre, the stop codon is excised, and gene expression proceeds. One popular lox-stop-lox plasmid is from Tyler Jack's lab: Lox-Stop-Lox TOPO.
- Cre-dependent gene knockout – conversely, putting the loxP sites on either side of a gene (called “floxing”, for “flanked by loxP”), will permit gene expression until Cre is present, at which time the gene will be disrupted or deleted.
- Selection marker removal – in conventional mouse targeting, targeted clones are selected for using a resistance marker; however, it is often desirable to remove the marker after the initial selection process. By floxing the selection marker, Cre can be used to easily perform this eviction.
- Regulated Cre expression – placing Cre downstream of promoters that are active only in certain cell or tissue types, during certain stages of development, or by making the Cre inducible (such as with tamoxifen or doxycycline), the Cre recombinase can be expressed only in specified cells or at specified times. Combining this with some of the loxP methods described above, a genetic modification can be restricted based on experimental constraints. This has been used for a wide range of purposes, including activating an oncogene only in a particular organ, or bypassing embryonic lethality.
To facilitate the use of Cre-lox technology, transgenic mice have been constructed that express Cre under a variety of ubiquitous and regulated promoters, and many loxP-containing alleles have also been constructed. Additionally, Cre-containing adenovirus (Ad-Cre) or AAV (AAV-pgk-Cre) has been used to successfully introduce Cre into cells of interest.
What if I need two separate recombination events?
One potential limitation of using loxP sites is the inability to tightly control which loxP sites recombine if more than two are present; intramolecular events happen with greater frequency than intermolecular events, but any two sites can potentially recombine. To account for this, alternate mutant versions of the loxP site have been created, which contain a unique asymmetric spacer "NNNTANNN", where "N" indicates which bases may vary from the canonical sequence. Among these are loxN (GtATACcT), lox2272 (GgATACtT), and lox511 (GtATACAT). These variant lox sites undergo recombination with other sites of the same type, but are not cross-compatible. Using different lox site variants allows Cre to catalyze more than one specific recombination event in a single system.
The Saccharomyces cerevisiae FLP-FRT recombination system is another site-directed recombination technology very conceptually similar to Cre-lox, with flippase (Flp) and the short flippase recognition target (FRT) site being analogous to Cre and loxP, respectively. The FLP-FRT technology can be an effective alternative to Cre-lox, and has also been used in conjunction with it, allowing for two separate recombination events to be controlled in parallel.
Have you used the Cre-lox system in your research? Do you have any tips or tricks, or clever cre-dependent techniques? Let us know in the comments!
References
1. Bacteriophage P1 site-specific recombination. I. Recombination between loxP sites. Sternberg, N. and Hamilton, D. 1981. PubMed PMID: 6276558.
2. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Sauer, B. and Henderson, N. 1988. PubMed PMID: 2839833. PubMed Central PMCID: PMC281709.
3. Tissue– and site–specific recombination in transgenic mice. Orban, P.C., Chui, D., and Marth, J.D. 1992. PubMed PMID: 1495975. PubMed Central PMCID: PMC49604.
4. Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre–loxP–mediated gene targeting. Gu, H., Zou, Y.R., and Rajewsky, K. 1993. PubMed PMID: 8513499.
Resources on Addgene.org
- Find Cre plasmids
- Find loxP plasmids
Topics: Plasmid Elements, Plasmids 101, Cre-lox, Plasmids







Leave a Comment