This post was originally written by Tyler Ford in 2018. It was updated by guest blogger Abhi Aggarwal in 2022.
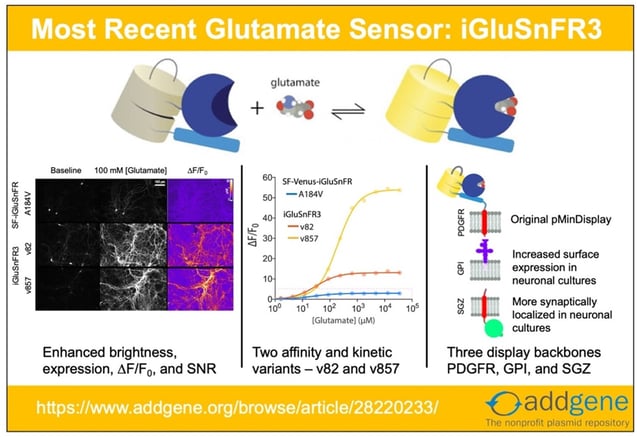
Recent updates to iGluSnFR and SF-iGluSnFR have made it clear that it’s time to update our iGluSnFR post! Here, we look at the origins of the system and explore advances that have happened since our original 2018 blog post.
The origins of iGluSnFR
iGluSnFR was developed in 2013 by Loren Looger’s lab at HHMI Janelia Research Campus to give researchers the ability to monitor glutamate release from neurons and other brain cells in vivo. Glutamate plays a variety of roles in synaptic communication and can trigger other forms of neuronal signaling and regulation.
To create iGluSnFR, the Looger lab wedged GFP between two halves of a bacterially derived periplasmic glutamate binding protein known as GltI. When the resulting fusion protein binds to glutamate, conformational changes lead to an increase in fluorescence intensity from the inserted GFP. After characterizing iGluSnFR in vitro, showing that it is selectively activated by glutamate, and tweaking it to improve its responsiveness, they showed that iGluSnFR could detect neuronal activity in C. elegans, zebrafish, and mice.
Second Generation iGluSnFR: SF-iGluSnFR
In the second generation of iGluSnFR, the Looger lab swapped eGFP for sfGFP to create SF-iGluSnFR. This improved variant has higher expression levels in bacteria and produces stronger fluorescent signals upon sensing glutamate in mouse and ferret models. Furthermore, they created three affinity variants – A184S (increased affinity for glutamate), S72A (decreased affinity for glutamate) and A184V (medium affinity for glutamate), that came in three different colors – blue (SF-Azurite-iGluSnFR), green (SF-iGluSnFR), and yellow (SF-Venus-iGluSnFR).
iGluSnFR3: Improved Glutamate Indicators for Synaptic Imaging
However, the first two generations of iGluSnFR had limitations: they were not optimal for experiments requiring high signal to noise ratios, imaging of high frequency glutamate signaling events, and accurately differentiating between synaptic vs. extrasynaptic glutamate release. Building on the previous generation of the SF-Venus-iGluSnFR (itself an improvement over iGluSnFR), Kaspar Podgorski’s lab have recently made iGluSnFR3 variants that are more sensitive and better localized for synaptic glutamate response than its predecessors. The detailed characterization of the new variants is available here.
Upgrades to iGluSnFR3
To improve upon the SF-iGluSnFR, the researchers did the following:
- Improve sensor’s photophysical properties such as kinetics, ∆F/F0, SNR, dynamic range, photostability, quantum yield, and expression.
- Create yellow (v82) and lime green (v857) variants differing in kinetics, affinity, and ∆F/F0.
- Introduce the two variants in three different membrane-targeting flavors – PDGFR, GPI, and SGZ, to improve neuronal membrane trafficking and localization.
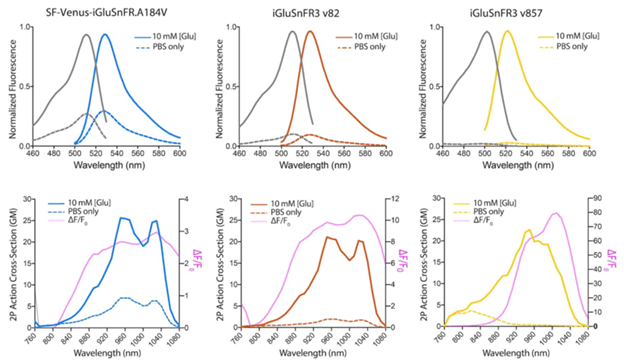
Improved photophysical properties
The improved variants of iGluSnFR3 have higher expression levels and produce stronger fluorescent signals upon sensing glutamate using both 1-photon and 2-photon imaging.
 |
|
Figure 1: 1P and 2P excitation and emission spectra of SF-Venus-iGluSnFR.A184V, iGluSnFR3 v82, and iGluSnFR3 v857. Image adapted from Aggarwal et al., Figure 1 |
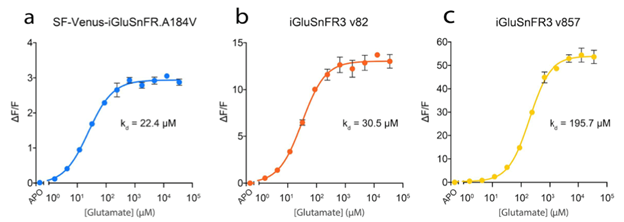
Increased versatility through two different variants
The yellow (v82) and lime green (v857) versions can be summarized quickly as:
- iGluSnFR3 v82: increased affinity and slower kinetics
- iGluSnFR3 v857: decreased affinity and faster kinetics
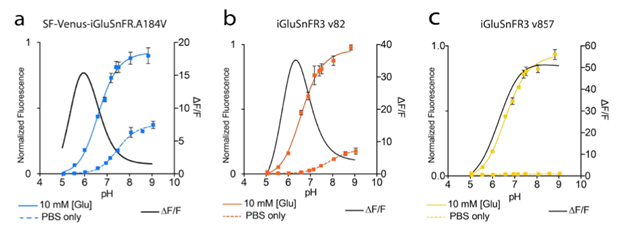
which are illustrated in figures 2 and 3.
 |
|
Figure 2: Glutamate titration curves for the different iGluSnFR variants. Image adapted from Aggarwal et al…, Supplementary Figure 3. |
 |
|
Figure 3: pH titration curves and ∆F/F0 for the different iGluSnFR variants. Image adapted from Aggarwal et al…, Supplementary Figure 2. |
Improved trafficking and localization through different C-terminal anchoring domains
The original iGluSnFR in the pMinDisplay backbone is fused to a specific peptide segment (PDGFR TMD) that anchors the sensor to the membrane and displays it on the extracellular side. In addition to the original PDGFR TMD, the researchers introduce iGluSnFR3 in two additional anchoring domains – GPI and SGZ.
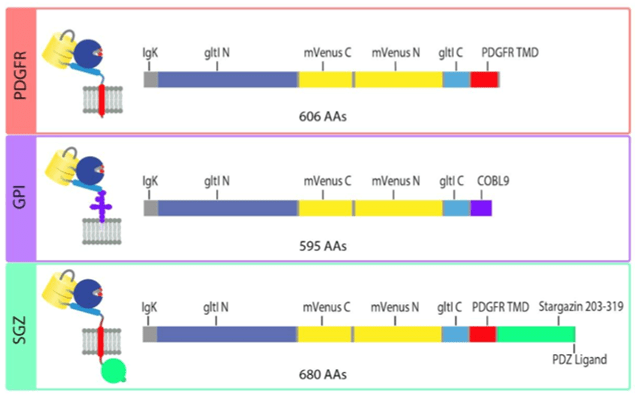 |
|
Fig: 4 Sequence schematics for PDGFR, GPI, and SGZ display constructs. Image adapted from Aggarwal et al… Figure 4. PDGFR is a C-terminal fusion to the PDGFR transmembrane domain in the mammalian expression pMinDisplay vector. GPI contains a C-terminal glycosylphostidylinositol anchor. SGZ contains a PDGFR transmembrane domain, followed by a modified form of the cytosolic C-terminal domain of Stargazin including a terminal PDZ ligand. |
The researchers recommend using the SGZ for in-culture work only, GPI anchors for in-culture work and 2P in vivo imaging, PDGFR for bulk 1P imaging or in new preps.
Using iGluSnFR3 in your lab
The new iGluSnFR3 variants should enable scientists to robustly detect signaling events in the brain and beyond.
These new iGluSnFR3 variants come in AAV packaged virus aliquots being deposited at Addgene. Other plasmids and pAAVs can be ordered directly from Addgene. The Podgorski Lab has provided a brief breakdown of the promoter systems you can use to express the iGluSnFR3 variants:
- pRSET: Bacterial expression vector, use with IPTG induction to express soluble protein
- CAG: Generic strong promoter. Formulated as AAV-DJ for neuronal expression.
- CAG-FLEX: CAG promoter, FLEXed for expression in Cre-expressing cells
- hSynap: Human Synapsin-1 promoter. Good for neuronal expression.
- hSynap-FLEX: Synapsin-1 promoter, FLEXed for expression in Cre-expressing cells.
- hSynap-FLP: Synapsin-1 promoter, FLP-dependent expression
- GFAP: Glial fibrillary acidic protein (promoter). Good for Glial expression (not neurons).
iGluSnFR3 is also available in several ready-to-use viral vectors.
References and Resources
Additional Resources on the Addgene Blog
- Learn about other Fluorescent Biosensors
- A Practical Approach to Choosing the B(right)est Fluorescent Protein
- Visit the Fluorescent Protein Featured Topic Page
Resources at Addgene.org
- Find Fluorescent Protein Biosensors
- Browse All Fluorescent Proteins
- Find AAV Tools
References
Marvin, Jonathan S., et al. “An optimized fluorescent probe for visualizing glutamate neurotransmission.” Nature Methods (2013) PubMed PMID: 23314171. PubMed Central PMCID: PMC4469972
Marvin, Jonathan S., et al. “Stability, affinity and chromatic variants of the glutamate sensor iGluSnFR.” Nature Methods (2018) PubMed PMID: 30377363
Aggarwal, Abhi, et al. . “Glutamate indicators with improved activation kinetics and localiation for imaging synaptic transmission” bioRxiv (2022)
Topics: Viral Vectors, Neuroscience Biosensors, AAV, Neuroscience







Leave a Comment