The hardest part of any revolutionary discovery, including CRISPR, is turning potential into impact. In molecular and cellular biology, this happens through the development of tools that exploit and expand our current knowledge. The Abudayyeh-Gootenberg lab (also called the AbuGoot lab), a joint lab run by Omar Abudayyeh, PhD, and Jonathan Gootenberg, PhD, focuses exclusively on this space — developing tools for cellular targeting. Lately, they’ve been working on two tools that expand and exploit CRISPR, both of which we're featuring in today's blog.
Cas7-11
Development and Discovery
As part of their quest to find, characterize, and develop new CRISPR systems and technologies, the AbuGoot lab often tests samples of new bacteria from anywhere they can find, whether the samples are from a rain gutter or the bottom of Tokyo Bay. When a weird, sulfurous smelling, black pellet was sent to the AbuGoot lab for testing, they weren’t sure what they would find… but it was safe to say the sample had their attention! They isolated DNA from the sample, amplified the CRISPR locus from the bacterial genome, and inserted it into E. coli to express and then test for any proteins that could cut RNA or DNA via a CRISPR mechanism. The weird black pellet's DNA yielded a new CRISPR effector protein they named Cas7-11.
Testing showed that Cas7-11 cut RNA with unprecedented specificity: when purified, it could be programmed to generate specific cuts on the RNA target. In addition, when they looked at the transformed E. coli expressing Cas7-11, they found no evidence of growth inhibition when the system was retargeted. This finding was surprising – in the most common RNA targeting system, Cas13, the "collateral" activity of the enzyme can lead to off targets and result in significant, negative effects in RNA targeting experiments. If Cas7-11 lacked this toxicity, it would represent a major step forward for CRISPR-based RNA editing - and further experiments strongly supported their initial findings.
The team originally suspected that this new Cas7-11 might be phylogenetically classified as the more complex Class I systems that contain multiple subunits, but the more they worked with it, the more they found evidence contradicting their initial assumption. Eugene Koonin and Kira Makarova, longtime collaborators of Omar and Jonathan and experts in CRISPR evolution, were also working on the project. Using computational observations, Koonin and Makarova found that Cas7-11 appeared to have derived from the more complex type III-A and III-B systems. Though it was composed of many protein subunits, it was nonetheless a new subtype.
Cas7-11 had combined all the domains into a single protein, shedding the complexity of its ancestors. They classified the system as Class I, Type III-E, and then used this information to identify a large variety of related bacteria with similar Cas systems. Some of these Cas systems have enzymes small enough to fit into a dual AAV system, albeit with lower knockdown efficiencies. The ability to identify related Cas systems opens up the potential of further development of low-toxicity RNA editing tools.
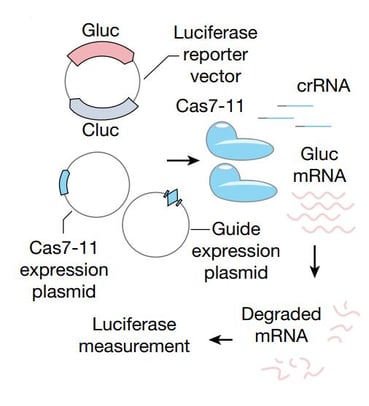 |
| Fig. 1 Schematic of luciferase RNA knockdown assays using Cas7-11 |
Use
The Cas7-11 targeting system contains two pieces: a single protein, computationally described as a fusion of four Cas7 proteins and a putative Cas11-like protein, and a guide RNA. The enzyme has dual activities: processing pre-CRISPR RNA into mature CRISPR RNA and cleaving RNA at positions defined by the target:spacer duplex without detectable non-specific activity. Its major advantage is the significantly reduced amount of off-target toxicity compared to other RNA editing methods — streamlining experimental design and its potential for clinical applications.
Specifically, efficacy rates of Cas7-11 were reported as up to 85%, broadly similar to shRNA and Cas13 systems in a range of mammalian cell lines. However, in those same comparisons, the AbuGoot lab reported no detectable effect on cell viability – meaning there was no measurable cell toxicity – using Cas7-11. This included tests done in cell lines known to be more sensitive to toxicity from RNA editing, such as mouse embryonic stem cells and glioblastoma cells.
PASTE
Development and discovery
The first generation of CRISPR could cut DNA, the second generation could do base editing, and with the third generation came Prime editing — the ability to now make any small changes, including base edits, insertions, and deletions. But CRISPR’s uses were still limited to small deletions and small insertions, leaving a several kilobase-sized gap in the gene editing field. The AbuGoot lab has now developed PASTE, which can “drag and drop” kilobases’ worth of DNA with CRISPR-driven precision into a genome, can be considered the fourth generation of CRISPR, one which drops itself neatly into that gap.
Consider, for instance, a disease like cystic fibrosis, which can be caused by thousands of different mutations, all found in the same gene. If we only have small, directed editing abilities, any gene editing treatment would have to be individually customized to the patient's specific mutations. With a drag and drop approach, however, the specific mutations wouldn’t matter – the mutated copy could be replaced by simply dragging and dropping in a functional copy of the entire gene, allowing a one-size-fits-all approach to gene replacement therapy for most patients.
But how does one go from “cut and paste” to “drag and drop”? The AbuGoot lab realized that they could likely do it using Prime editing alongside some creative (and significant!) protein engineering. They combined Prime editing with an integrase via protein fusions, using Prime to lay a “beacon” into the genome that a fused integrase would then integrate into with a large DNA insert – or Programmable Addition via Site-specific Targeting Elements (PASTE).
Finding the right integrase for this new approach proved a challenge. The old standby of CRE/flp didn’t work – it turns out these tyrosine recombinases are as bad at integrating as they are good at flipping. So the lab instead turned to phage serine integrases, where they saw 10-20% integration of inserts up to 36 kb into a genome. This integration was observed in non-dividing cells, meaning that this system wasn’t dependent on the cell’s intrinsic machinery being already activated to work. (It was also found to be independent of gene repair.) This lowers the chance of the insert being “dropped” in the wrong place, while increasing the number of potential applications.
But they weren’t done engineering quite yet – surely, they thought, the integration rate could be improved upon. The lab switched up the serine integrase for one which can enzymatically take a genetic sequence and integrate it into any DNA sequence containing the 38-50 bp sequence recognized by the serine integrase, finding tens of thousands new integrases with efficient integration activity in human cells. Moreover, they optimized the specific attachment sites via sequence engineering, enabling even more efficient integration activity. They now had a system that could integrate genes up to 36 kb in length, with an integration rate of 10-55% - the PASTE system.
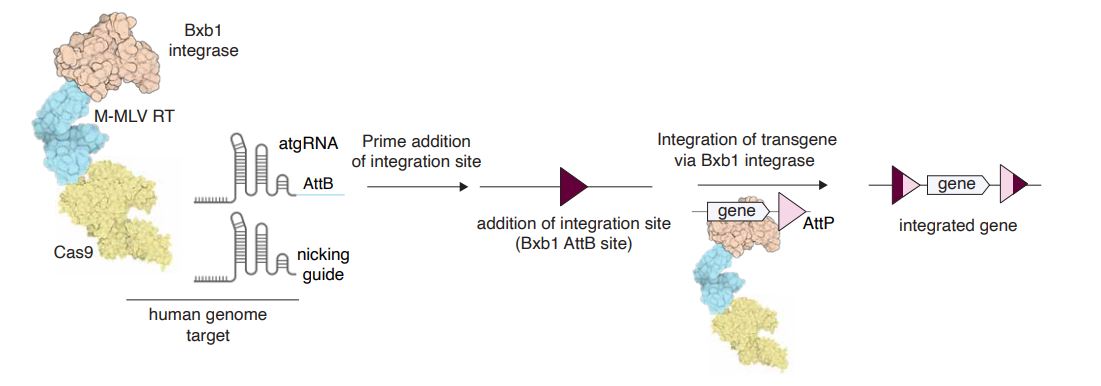
|
| Fig. 2: Schematic of programmable gene insertion with PASTE. The PASTE system involves insertion of landing sites via Cas9-directed reverse transcriptases, followed by landing site recognition and integration of cargo via Cas9-directed integrases. |
Use
PASTE can be delivered via a single dose of plasmids, works in non-dividing and primary cells, and can be easily programmed for new genes. In comparison with other large insert techniques like HITI or HDR, which require specific repair pathways to work, PASTE is more specific and efficient than HITI and has higher purity of insertion than both HITI and HDR. PASTE is also compatible with both AAV and adenovirus templates.
The system, which is larger than Cas9, can also be used with mRNA and a synthetic guide. Like PRIME, PASTE relies on single-stranded nicks instead of the double-stranded breaks used in other insertion systems. Off-target nicks are much less harmful than off-target breaks, almost always resolved without any toxic effects to the cell. This means almost no indels and no off-target effects, giving PASTE a high-fidelity rate.

Both Cas7-11 and PASTE open up multitudes of possibilities at the bench and potentially in therapeutic applications. The potential of CRISPR has yet to fully taken advantage of, but exciting new tools like these represent a major step forward for what the field can do.
References and Resources
References
Özcan A, Krajeski R, Ioannidi E, Lee B, Gardner A, Makarova KS, Koonin EV, Abudayyeh OO, Gootenberg JS. Programmable RNA targeting with the single-protein CRISPR effector Cas7-11. Nature. 2021 Sep;597(7878):720-725. doi: 10.1038/s41586-021-03886-5. Epub 2021 Sep 6. PMID: 34489594.
Eleonora I. Ioannidi, Matthew T. N. Yarnall, Cian Schmitt-Ulms, Rohan N. Krajeski, Justin Lim, Lukas Villiger, Wenyuan Zhou, Kaiyi Jiang, Nathaniel Roberts, Liyang Zhang, Christopher A. Vakulskas, John A. Walker II, Anastasia P. Kadina, Adrianna E. Zepeda, Kevin Holden, Jonathan S. Gootenberg, Omar O. Abudayyeh. Drag-and-drop genome insertion without DNA cleavage with CRISPR-directed integrases bioRxiv 2021. 11.01.466786; doi: https://doi.org/10.1101/2021.11.01.466786
Additional Resources on the Addgene Blog
Prime Editing: Adding Precision and Flexibility to CRISPR Engineering
CRISPR 101: RNA Editing With Cas13
Resources at Addgene.org
Topics: CRISPR, Other CRISPR Tools






Leave a Comment