The central nervous system (CNS) orchestrates complex processes enabling organisms to control their movements and behavior. These functions and others are controlled by collections of neurons that are intricately wired into circuits through synaptic connections (Shepherd, 2004). Understanding the structure and function of these neural circuits is essential for neuroscience research. The use of genetic tools for visualizing and perturbing circuits together with the development of methodologies to deliver genes to the CNS have recently made it much easier to map these neuronal networks.
For almost three decades, recombinant Adeno-associated virus (AAV) has been widely used to deliver genetic tools to the brain because of its low cytotoxicity, low immunogenicity, and the long-term expression of AAV delivered genes. AAV serotypes have been extensively studied in order to define those that target specific neuronal populations in the brain. The Allen Institute and the Janelia Research Campus in the USA have used AAV1 and AAV2 serotypes in order to create an atlas of the mouse brain. This atlas enables researchers to quickly find the best AAV serotypes to target the neurons they’re studying. Thanks to this work it is now much easier to express molecules in your favorite neuronal population and track connections to that population.
Limitations of common genetically encoded neuronal tracers
Neuronal tracers such as epitope tags and fluorescent proteins (FP) have also been important in mapping, monitoring, and manipulating neuronal circuits. Epitope tags are short antigenic peptide sequences, attached to a protein of interest (POI), that facilitate immunohistochemistry (IHC) experiments with tag-specific antibodies. The most famous ones are influenza hemagglutinin (HA), myelocytomatosis viral oncogene (myc), simian virus 5-derived epitope (V5), the synthetic peptide FLAG, the synthetic streptavidin-binding strep-tag and, more recently, OLLAS (Escherichia coli OmpF linker and mouse langerin) and Sun Tag (Viswanathan, 2015).
The principal advantage of epitope tags for IHC is the availability of reliable primary antibodies for their detection, particularly when antibodies to the POI are non-specific, cross-react with other antibodies, or are unavailable entirely. The other advantage is that these tags are small (8-12 amino acids) and neither disturb POI folding nor its ability to interact with protein partners within the cell. However, antibodies can have low affinity for these tags and even multimeric tags are frequently insufficient for detection when the POI is weakly expressed. As these epitopes tags have no intrinsic visual signal, they cannot be detected directly in live imaging experiments. It is therefore difficult to use epitope tags to monitor single molecules - something researchers often want to do at neuronal synapses for instance. Also these tags need to be fused to scaffold proteins in order to be expressed stably in neurons.
As compared to epitopes tags, FPs are intrinsically fluorescent and can therefore be used in live imaging experiments crucial to studying neuronal circuits and dynamics. FPs are generally bright, stable and well tolerated by cells. They can be used for protein localization, isolation, and tracking experiments (Rizzo, 2009). With many different FPs that emit across the visible spectrum, researchers can also use multiple FPs in single experiments and visualize numerous molecules or structures at once (Shaner, 2007). In contrast to epitope tags, FPs are bigger and their fusion to a POI can affect protein folding and protein-protein interactions. Also, overexpression of FPs can trigger protein aggregation which is toxic for the cells and affects the labeling of small structures like neurites. Like epitope tags, low FP expression may be insufficient for monitoring a weakly expressed POI.
Spaghetti Monster - A new and improved neuronal tracer
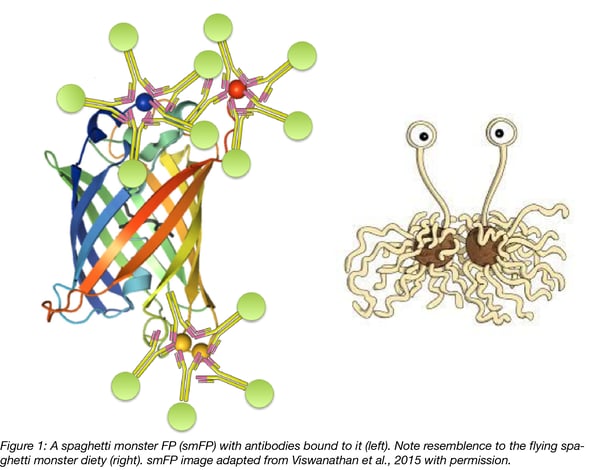
With the limitations of epitope tags and fluorescent proteins, it has been crucial to develop new tracers that enable accurate labeling of neurons. As discussed above, weakly expressed proteins in particular are difficult to track with these classic probes. To overcome the limitations of existing FP and epitope tags, researchers in Loren Looger’s lab have developed new molecular tags combining the advantages of both (Viswanathan, 2015). For Looger’s team, an ideal probe should combine the solubility, cell tolerance, and optional fluorescence of FPs with facile antibody recognition of epitope tags. Thus they have created antigenic protein tags called “spaghetti monster” fluorescent proteins (smFPs). smFPs are literally composed of FPs fused to multiple epitope tags. Indeed, to create the smFPs Looger’s team strategically inserted 10 to 15 copies of single epitope tags into single FP scaffolds with either intact or darkened chromophores. These probes are both intrinsically fluorescent and can be detected with highly epitope-specific, fluorescently-labeled antibodies.
The “spaghetti monster” name is a reference to the deity of the social movement, the church of the Flying Spaghetti Monster; the smFP structure resembles images of the deity (see comparison image above). The church of the Flying Spaghetti Monster is a symbol of opposition to the teaching of intelligent design in public schools and was started in 2005 in Kansas by Bobby Henderson, a physics graduate from Oregon State University.
Find the Spaghetti Monster probes at Addgene
Uses of smFPs
1. Multi-channel connectomic tracers
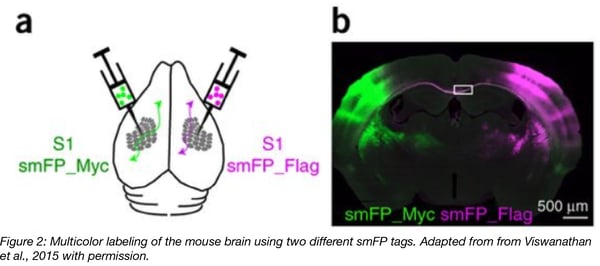
Fluorescent proteins such as GFP have been used for a long time as probes to study connectomics (how neural circuits are organized and how neurons connect to each other). Combined with AAVs, FPs can be easily expressed in the brain and can be detected through their direct fluorescent signals or using anti-FP antibodies.
GFP emits light in only a single channel. In multi-color imaging experiments, the second color is usually provided by red fluorescent proteins such as tdTomato or RFP or blue fluorescent proteins like Cyan. The issue with these FPs is that they are less photostable than GFP. In addition, antibody-based amplification of their signals is weak and increases background fluorescence. Furthermore, many of these FPs are cytotoxic, prone to aggregation and cross-react with anti-GFP antibodies, making them difficult to use in a multicolor/multichannel imaging set-up. In contrast, smFP tags can be recognized by distinct, fluorophore-labeled antibodies with very specific reactivities making them more useful for multicolor imaging experiments than standard FPs. The Looger Lab was thus able to easily perform a four-color labeling experiment using smFPs and standard neuronal tracers.
2. Visualization of neuronal sub-cellular structures
GFP is limited in its ability to label subcellular structures in the CNS. GFP and other FPs have the tendency to brightly label somata but not neuronal processes. In contrast, smFPs can label fine neuronal structures with high fidelity and at lower concentrations than GFP. smFPs enable better labeling of these fine structures through their multiple, high-affinity binding sites for primary antibodies which can be used to amplify the direct smFP signal within sub-cellular structures. For instance, smFPs have been used to visualize “thorny excrescence” (TE) spines in the Hippocampal CA3 pyramidal neurons which are known to be notoriously difficult to label. It has also been shown that dendrites in the stratum oriens and the stratum lacunosum moleculare were better resolved with smFP_FLAG than GFP.
3. Enhanced ability to distinguish weakly expressed and/or similar proteins
Through their multiple, highly specific antibody binding sites and primary antibody-based detection, smFPs create much brighter signals for weakly expressed proteins than standard tags and can be used to distinguish highly similar proteins. For example, N-cadherin (cadherin-2) is a postsynaptic cell adhesion protein that plays a critical role in neural development. It belongs to a huge family of proteins that are highly similar in terms of sequence and thus very difficult to distinguish using standard antibodies. By fusing N-cadherin to smFPs the Looger team was able to specifically label N-cadherin in neurons and show that smFPs fusions provided better labels than fusions to 3 or more HA tags.
4. High-resolutions microscopy
As indicated above, smFPs can be used with conventional microscopes, such as epifluorescence or confocal microscopes, but they can also be used with the most cutting-edge imaging technologies such as array tomography, super-resolution STORM imaging, and electron microscopy. Sample preparation for these techniques sometimes makes it more difficult to detect protein tags using antibodies. In contrast, with their many epitopes, smFPs retain their ability to be detected after sample preparation better than standard proteins tags like GFP. Specifically for STORM, the high intensity signal provided by smFPs can result in better resolution images.
Conclusion
By combining advantages of FPs and epitope tags, smFPs appear to be very effective probes for finely visualising sub-cellular structures or low-abundance proteins. When delivered with AAV, these probes can be used to label neuronal circuits and structures that are usually difficult to see. smFPs are an excellent add-on to the neuroscientist’s tool-kit for studying the brain and we looked forward to the expanded use of smFPs in the future.
References
1. Viswanathan, Sarada, et al. "High-performance probes for light and electron microscopy." Nature methods 12.6 (2015): 568. PubMed PMID: 25915120. PubMed Central PMCID: PMC4573404.
2. Shepherd, G.M. (2004). In "The synaptic organization of the brain". Oxford University Press, New York. pp 719.
3.Rizzo, Mark A., Michael W. Davidson, and David W. Piston. "Fluorescent protein tracking and detection: fluorescent protein structure and color variants." Cold Spring Harbor Protocols 2009.12 (2009): pdb-top63. PubMed PMID: 20150100.
4. Shaner, Nathan C., George H. Patterson, and Michael W. Davidson. "Advances in fluorescent protein technology." Journal of cell science 120.24 (2007): 4247-4260. PubMed PMID: 18057027.
Additional Resources on the Addgene Blog
- Rabies and neuronal tracing
- Retrograde AAV: Making the journey from axon to nucleus
- AAVs CREATEd for gene delivery to the CNS and PNS
Resources on Addgene.org
- Find the Spaghetti Monster probes at Addgene
- Browse all AAV vectors
- Find ready-to-use AAV preps
Topics: Viral Vectors, Cell Tracing, Neuroscience







Leave a Comment