In the short time since its development, CRISPR/Cas9 genome editing has been used to study the effect of gene knockout in vivo and in vitro, as well as to insert targeted mutations through homologous recombination. To further increase the utility of CRISPR/Cas9, it will be necessary to improve its multiplexing capacity. Multiplexing is key due to the natural redundancy of biological pathways; to observe a phenotype, the modification of multiple genes is often necessary.
Guide RNAs (gRNAs) are commonly packaged in 400-500 bp cassettes containing the RNA pol III promoter, gRNA and pol III terminator. These relatively large cassettes (considering the gRNA itself is ~100 bases) limit the number of gRNAs that can be packaged together in a single vector. In addition, the pol III promoter is relatively weak, and low expression of gRNAs from these constructs could lower genome editing efficiency.
One previous strategy for multiplexing gRNAs used the RNA nuclease Csy4 from Pseudomonas aeruginosa to cleave 4 gRNAs separated by 28 base Csy4 recognition sites from a polycistronic transcript. Although this method shortens construct size, it requires coexpression of Csy4 with Cas9 and the polycistronic transcript. As nucleases may have off-target effects and/or toxicity, it would be advantageous to instead use a preexisting, conserved cellular machinery to process gRNAs.
Hijacking tRNA processing to make gRNAs
In a recently published paper, Yinong Yang’s group did just that as part of their CRISPR work in rice plants. Yang was intrigued by the tRNA processing machinery present in almost every living organism. Precursor, or pre-tRNAs are cleaved by RNase P and Z in eukaryotes (E in bacteria), and only a few elements of tRNAs are necessary for recognition and cleavage. This sequence flexibility indicates that gRNAs could be incorporated into these constructs without compromising cleavage efficiency. In addition, tRNAs contain internal transcriptional elements that enhance pol III transcription, potentially increasing the levels of gRNA expression. Since tRNAs are among the most abundant molecules in a cell, this system is clearly also robust enough to ensure high levels of gRNA cleavage.
Xie et. al constructed polycistronic glycine tRNA-gRNA genes (PTGs) in the forms tRNA-gRNA and tRNA-gRNA-tRNA, with short 5’ spacer sequences preceding gRNAs, to test for gRNA cleavage. gRNAs were cleaved precisely as predicted with no addition of nucleotides to the 5’ spacers. The 3’ ends were modified slightly with either a 1-7 base poly(U)tail in tRNA-gRNA constructs or a 1-4 base tail from the second tRNA in the tRNA-gRNA-tRNA constructs. gRNAs with any 5’ leading nucleotide could be processed, a distinct advantage over other methods that favor certain nucleotides at the 5’ end.
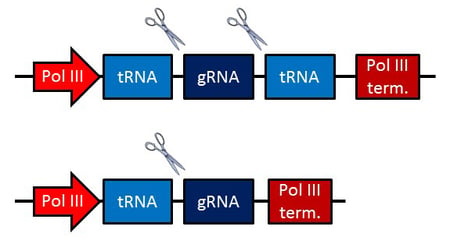
PTG construct schematics. Scissors indicate cleavage sites.
After confirming gRNA cleavage, they examined gRNA expression levels and indel generation for two PTG constructs in rice protoplasts. Compared to matching traditional gRNA-expressing constructs, transcript levels of PTG1 and PTG2 were 3 and 31 times higher, respectively. In rice protoplasts, Xie et. al observed slightly higher mutation rates than they’d previously reported in protoplasts (15% and 9% of target sites.)
How many loci can Cas9 modify?
To test the application of this system to gRNA multiplexing, Xie et al. constructed PTGs containing 8 gRNAs targeting 4 MAPK genes, again using rice protoplasts. Each pair of gRNAs targeted genomic sites separated by 350-750 bp, allowing them to examine the frequency of both indels at each locus, as well as large deletions. They detected fragment deletion with 4-20% frequency at the four loci, indicating that Cas9 can be guided to at least 8 distinct loci.
They also examined editing efficiency in intact rice plants, comparing traditional gRNA constructs with PTGs. In PTG constructs containing two gRNAs, they found 100% indel frequency at both targets (35% and 76% biallelic), compared with only 44% and 66% indel frequency with the use of standard single gRNA constructs. When PTG constructs with five gRNAs were tested, they found that 50% of plants had biallelic mutations at all five loci. This highly efficient multiplexing further illustrates the potential of PTGs.
Applications of PTGs
The major advantage of the PTG system is its universality; robust and efficient tRNA processing is common to prokaryotes and eukaryotes. Though the plasmids made by Xie et. al are specifically designed for plant expression, PTGs will likely be adaptable to many different biological systems. Although Xie et. al characterized the system in plants, PTGs will likely work well in many different biological systems. Size is also a factor; the pol lll promoter and terminator need be encoded only once, and each <200 bp tRNA-gRNA takes up half the space of a standard pol III-gRNA, which will facilitate CRISPR/Cas9 multiplexing in systems with limited packaging capacity.
Still confused about the various tRNA expression strategies? No worries, I've summarized the three strategies described in this post in the graphic below - including some simple comparisons.
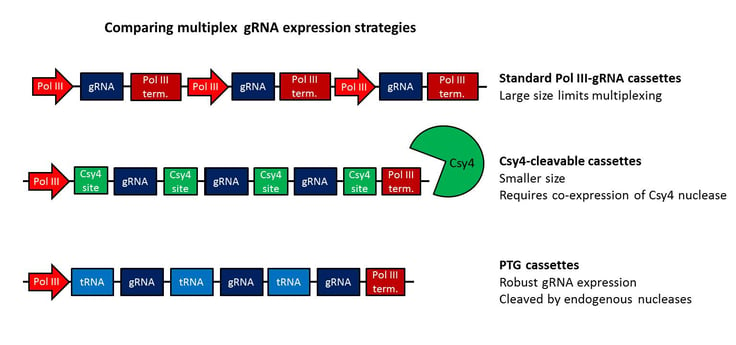
The easy multiplexing afforded by PTGs will improve the utility of CRISPR/Cas9 in new avenues of research. The use of multiple gRNAs targeting a single gene can increase the likelihood of gene knockout, or multiple gRNAs may be used to target genes with overlapping activity. Multiple gRNAs can also facilitate deletions of other regions, such as noncoding RNAs or regulatory elements, to examine their function. Bacteria and viruses have illustrated the power of polycistronic gene organization, and its translation to CRISPR/Cas9 shows great potential for multiplexed genome editing.
Interested in using PTG plasmids in your research? Check out the PTG plasmids or plant expression systems available from Addgene. If you have any questions or thoughts about this technology, leave them in the comments below.
References
- Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Xie K, Minkenberg B, Yang Y. Proc Natl Acad Sci U S A. 2015 Mar 17;112(11):3570-5. doi: 10.1073/pnas.1420294112. Epub 2015 Mar 2. PubMed.
- Find the plasmids from this publication at Addgene
Other Techniques for gRNA Multiplexing
- Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Nissim L, Perli SD, Fridkin A, Perez-Pinera P, Lu TK. Mol Cell. 2014 May 22;54(4):698-710. doi: 10.1016/j.molcel.2014.04.022. Epub 2014 May 15. PubMed.
- Find the plasmids from this publication at Addgene
- Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK. Nat Biotechnol. 2014 Jun;32(6):569-76. doi: 10.1038/nbt.2908. Epub 2014 Apr 25. PubMed.
.- Find the plasmids from this publication at Addgene
Topics: CRISPR, CRISPR gRNAs







Leave a Comment