Who doesn’t love a library?
A traditional library might contain thousands or millions of different books. Addgene’s pooled libraries are a little different: each is a collection of plasmids that share the same backbone and differ only in a small region. This makes them useful for a wide variety of high-throughput experiments.
There are currently 390 pooled libraries available at Addgene, with more added to the collection all the time! We’ll serve up a sampling of some of the most popular and give you a tour of what these libraries are capable of at the same time.
Barcoding libraries
Just as physical barcodes help librarians track individual books, genetic barcodes allow researchers to identify specific cells — as well as their descendants. This makes them useful for lineage tracing, among other applications. Cells can be identified using fluorescence, like in the pMuSIC library, or by sequencing the unique DNA barcodes.
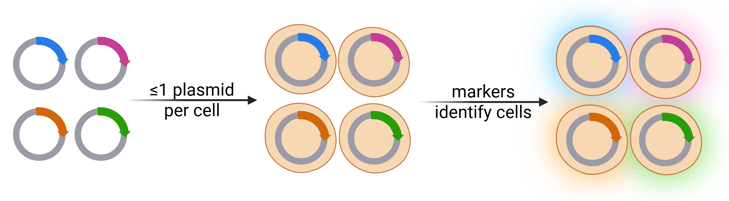 |
| Figure 1: Schematic of barcoding with a pooled library. Created with BioRender.com. |
The CellTag libraries from the Samantha Morris lab use a combinatorial barcoding approach to observe clonal dynamics and cell lineage. Variations of this library allow tracking of transcription or chromatin accessibility at the single-cell level.
CRISPR libraries
CRISPR screening has become a popular tool for probing cellular pathways and mechanisms. CRISPR screening libraries supply a collection of sgRNAs, but they may or may not include CRISPR machinery (which can be supplied separately). Libraries can target specific pathways or the entire genome at once!
Knockout screening with CRISPR libraries
In CRISPR knockout screening, sgRNAs guide Cas9 to cut targeted genes, creating mutations that prevent their expression. The knockout cells can be used to identify genes that underlie a particular phenotype.
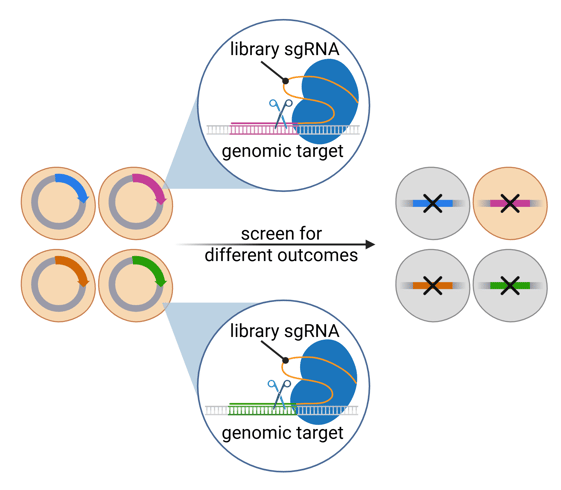 |
| Figure 2: Schematic of CRISPR knockout screening with a pooled library. Created with BioRender.com. |
The popular (and deliciously named) Brie and Brunello libraries have been used for hundreds of genome-wide knockout screens in mouse and human cells, respectively. Last month, the David Root and John Doench labs deposited new and improved libraries called Julianna, for mouse screening, and Jacquere, for human screening. These new libraries have improved on-target activity for more robust and accurate screens.
Activation or inhibition screening with CRISPR libraries
Libraries for activation or inhibition CRISPR screening are similar to those for knockout screens: sgRNAs to guide Cas9 to targeted genes. In these applications, however, nuclease-dead Cas9 is fused to a transcriptional activator or repressor to create a reversible effect. These approaches may also be less toxic than knockout screens.
The Jonathan Weissman lab has deposited genome-wide CRISPR activation (CRISPRa) libraries for both mouse and human use and CRISPR inhibition (CRISPRi) libraries for both mouse and human use. Each of these libraries accounts for chromatin, position, and sequence features in the selection of sgRNAs.
Other CRISPR libraries
As CRISPR technologies multiply, so do CRISPR pooled libraries! Addgene’s collection also includes a base editing sensor library, a base editing knockout library, a prime editing assessment library, and a prime editing lineage tracing library.
Other screening libraries
CRISPR isn’t the only game in town! Pooled libraries can be designed for all kinds of different high-throughput experiments. Our most popular screening library is the MORF library, which stands for Multiplexed Overexpression of Regulatory Factors. The Feng Zhang lab developed this library of all human transcription factors, including splice isoforms, to allow comprehensive screening, cellular programming, and more. For more details, check out our blog post on this library.
Let’s also highlight a prokaryotic library: the Cultivarium MACKEREL Library, whose acronym means Modular, NGS-trACKable ExpRession Element. This library consists of broad host range regulatory sequences ready to be assembled with other elements using golden gate cloning. This toolkit is designed to be used across many species of bacteria.
Surface display libraries
In surface display, proteins of interest are fused to anchoring proteins to be expressed on the surface of a host cell or virus, such as a yeast or bacteriophage species. This approach is useful for screening for binding properties. For example, several coronavirus libraries have been used to systematically explore variations in the spike protein, teaching us more about viral evolution and antibody binding.
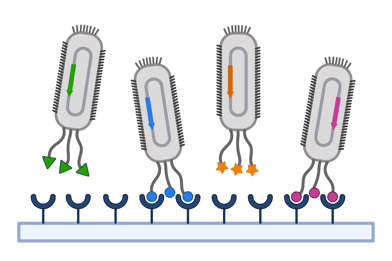 |
| Figure 3: Schematic of phage display screening with a pooled library. Surface display screening can also be conducted with yeast. Created with BioRender.com. |
Conclusion
Have we whetted your appetite for pooled libraries? Addgene’s pooled library collection page features hundreds more libraries to browse. Our Guide to Plasmid Pooled Libraries has tips on using these libraries and designing screens with them.
All pooled libraries are available as pooled plasmid DNA. In addition, several libraries are offered as lentiviral preparations to save you a few steps! To check whether a library you’re interested in is available as a viral prep, look under “Ordering” to see what options are offered.
 |
| Figure 4: Screenshot of the Addgene website. |
Let us know which pooled libraries you’ve used in the comments!
Resources
Additional resources on the Addgene blog
Resources on Addgene.org
Topics: CRISPR Pooled Libraries, Plasmids, lentivirus





Leave a Comment