Transcription factors (TFs) act as the gate keepers for gene expression, turning transcription on and off by binding proximal to their target genes. Since gene expression patterns determine everything from stem cell differentiation fate to tumor suppression (and most everything in between), understanding how, when, and where TFs work is of broad interest to the scientific community. Recently, the Zhang lab generated a TF library (aka the MORF Collection) and an accompanying expression atlas, including over 3,500 human TF isoforms, to help researchers understand this regulatory network.
The transcription atlas and library
The transcription factor library
The human genome encodes approximately 1,800 TFs, with >3,500 unique isoforms transcribed from these sites. The Zhang lab generated a comprehensive library of each unique TF open reading frame (ORF), barcoded for tractability. These ORFs are individually overexpressed and available for lentiviral packaging and delivery to target cells as a pooled library, which will amplify each TF to determine how its presence influences cell fates.
The atlas of directed differentiation
To understand how each human TF can contribute to differentiation, the Zhang lab transduced human embryonic stem cells with the viral TF library, with each cell receiving only 1 TF expression construct. Cells were cultured following a differentiation protocol before performing single-cell RNA sequencing. The sequencing results covered TFs representing 92% of the MORF library, with an average of 206 cells per TF ORF. From this large sequencing endeavor, a map of possible TF-induced cellular states was generated (Joung et al., 2023).
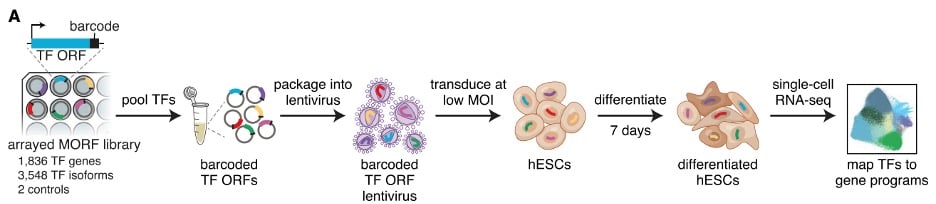 |
| Fig. 1: Barcoded TF library was packaged in lentiviral particles and used to transduce human embryonic stem cells. Transduced stem cells were differentiated and single cell RNA-seq was performed to map TFs to cell fates. (Adapted from Joung et al.) |
This atlas linked TF expression to differentiation outcomes as well as global transcriptional changes. Using this road map, and what was known about previously characterized TFs, the Zhang group were able to classify unknown, or ‘orphan’, TFs into their respective families. Given the number of poorly studied TFs, this functional association of the uncharacterized TFs is a useful finding.
Transcriptional program changes from this screen successfully differentiated cells resembling all three germ layers (ectoderm, endoderm, and mesoderm). These profiles were then compared to that of reference cell types generated by the human fetal expression atlas. Computational analysis was used to infer differentiation trajectories and identify both up- and down-regulated pathways. Individual cellular fates were then matched with candidate TFs responsible for that differentiation.
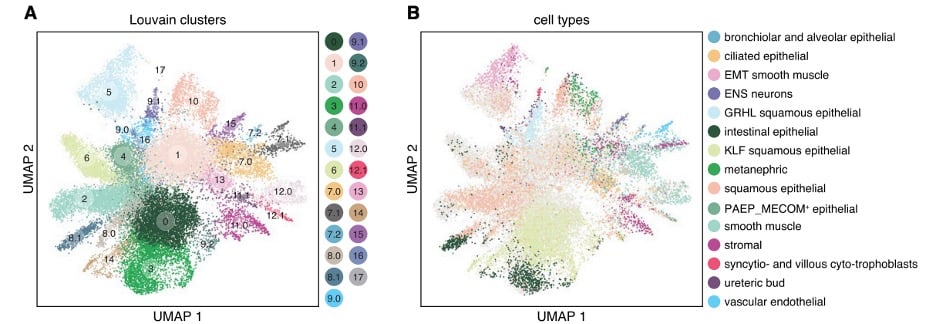 |
| Fig. 2: Single cell RNA-seq clustering of cells transduced with the TF pooled library. Louvain community clusters of individual cells along with color coded nominated reference cell types. (Adapted from Joung et al.) |
Putting MORF to work
Screens and selections
In its current format, the MORF library is most immediately useful for TF library screens. Previously engineered TF libraries were not as comprehensive as this one, so it is an invaluable tool to be used in this capacity. Similar to the Human TFome Library, the MORF library can be used with other selection and screening mechanisms, although the unique barcodes for each TF help streamline identification.
While the Zhang lab controlled their multiplicity of infection so that a single TF was delivered to each cell, future applications could deliver multiple TFs to each cell, allowing for combinatorial overexpression programs on cellular processes. For help with library transformation and amplification, refer to the Zhang lab’s previously published protocol and its relevant modifications on the pooled library webpage.
Disease models
Good disease models can often be difficult to generate, in part because of difficulty with replicating the specific dysregulation in the disease. For example, very few diseases manifest as a single mutation or expression dysregulation event. Diseases are often complex, with multiple layers of dysregulation from either genetic mutation and/or TF dysregulation. A targeted TF screening approach can not only lead to a better understanding of disease onset, but also better simulate a disease in a model system. TF screens can be designed to identify custom TF programs to acutely initiate disease onset, providing both a tool to test prophylactics on and a program that any researcher could use to generate the relevant disease model. Accurate modeling of a disease depends on both genetic and expression control systems to mimic illness on a molecular level.
Atlas resource
The TF map is a valuable resource for anyone interested in cell differentiation programs. The Zhang lab validated their Atlas’s TF fate map by selecting a panel of candidate TFs which were predicted to generate unique cell types. Of the 24 TFs they tested, 22 expressed markers consistent with the cell type that was predicted based on the Atlas. They also looked at 17 of these TFs to see if the differentiated cells had morphologies consistent with the predicted cell type (e.g., axon-like projections for neuron predicted differentiation), and 15/17 morphologically aligned. Documentation of the network that initiates biological reprogramming is a valuable reference if you are looking to understand a pathway, or just looking for the best astrocyte differentiation protocol!
Ready to try it out for yourself? Check out the MORF Collection here! The pooled library comes either with or without mCherry and GFP fluorescent controls. Alternatively, you can order individual plasmids from the library to further investigate specific genes and TF isoforms. Due to the size of the collection, we’re releasing individual plasmids in batches. If there’s something you’d like that’s not yet available, just ask an Addgene scientist at help@addgene.org!
References and Resources
References
Joung J. et al., A transcription factor atlas of directed differentiation. Cell. 2023. 186 (1) 209-229. doi: 10.1016/j.cell.2022.11.026
Joung J. et al., Genome-scale CRISPR Cas-9 knockout and transcriptional activation screening. Nat Protoc. 2017 (4):828-863. doi: 10.1038/nprot.2017.016.
Additional Resources on the Addgene Blog
Additional Resources on Addgene.org
- Plasmid Pooled Libraries
- Pooled library amplification
- Zhang Lab CRISPR Plasmids
- The Human TFome Library
Topics: Plasmids





Leave a Comment