If you take a look in a typical lab freezer, you’ll probably find lots of different plasmids containing the same protein of interest tagged with different fusion proteins for different experiments over the years — a green fluorescent protein, a red fluorescent protein, a fancy new green fluorescent protein, some affinity purification tags, another version with that same red fluorescent protein somebody made because they forgot it was already in the freezer… you get the idea. What if there were a way to just make one fusion protein that switches to any color or tag you want?
It sounds like magic, but it’s real! In this post, we’re going to talk about the power and flexibility of self-labeling protein tags.
The basics
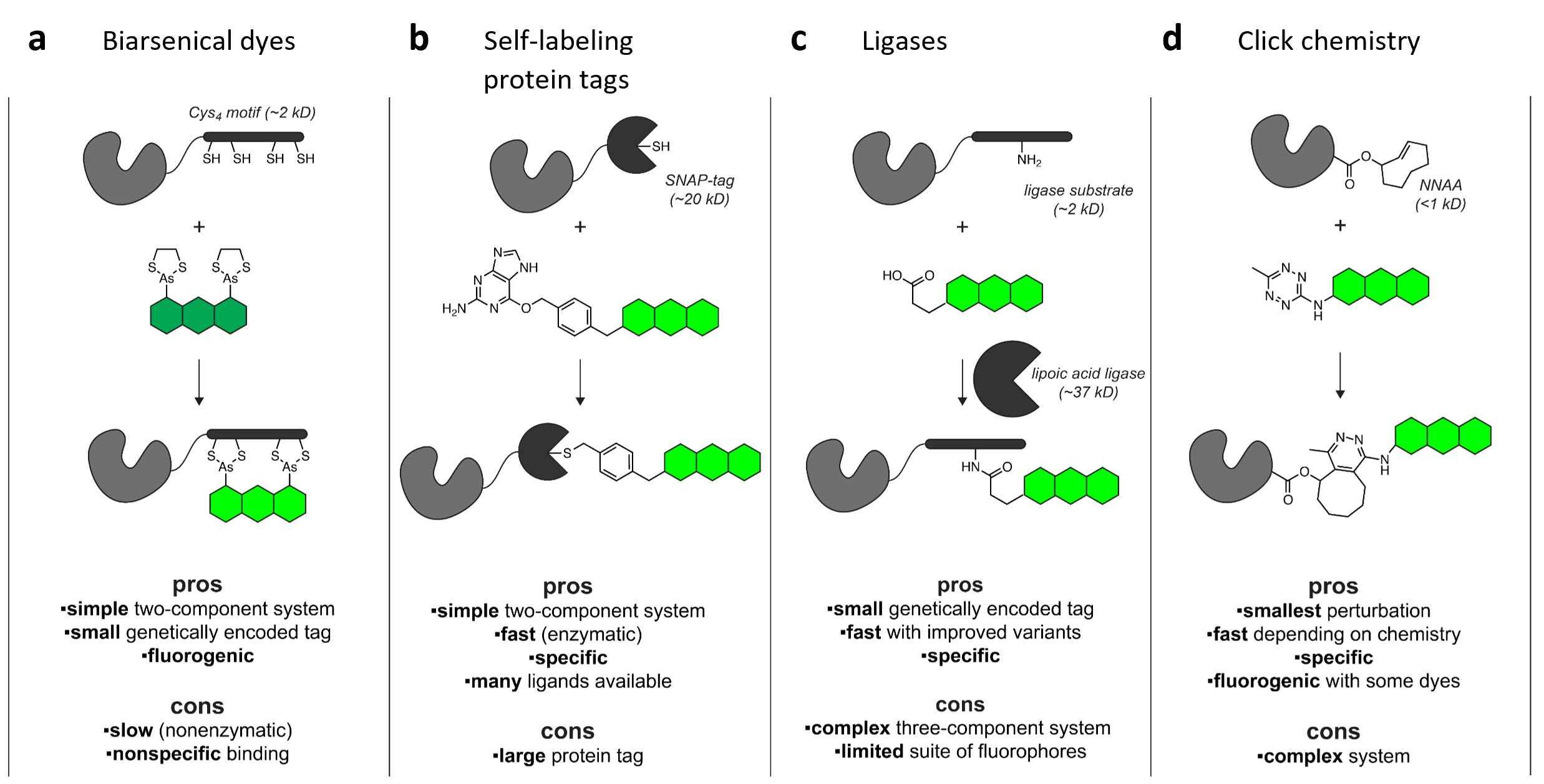
These tags most often consist of an enzyme that has been mutated to block its catalytic activity and instead become trapped in a covalent bond with its substrate. Then, all it takes is a bit of chemistry magic to produce a wide range of useful molecules linked to that substrate. If the substrate was linked to, say, a chemical dye or other functional ligand, your protein of interest will end up linked to whatever molecule you choose (Figure 1). Just express the fusion protein and add a specific ligand!
A self-labeling protein gives you the flexibility to choose different colors or ligands for different experiments without having to clone a whole new construct for every application. Plus, the photophysical properties of chemical dyes provide many advantages over fluorescent proteins, opening up new applications.
 |
|
Figure 1: Live-cell labeling strategies. The green structure represents a fluorophore or other useful ligand, gray structure is the protein of interest, and black is the fusion tag or labeling system. Image adapted from Lavis, 2016, under CC-BY. |
Common self-labeling tags
Let’s take a look at some popular self-labeling tools:
Tetracysteine: The first reported chemical labeling tag, from Roger Tsien’s lab in 1998 (Griffin et al., 1998), this 15 amino acid peptide contains a cluster of four cysteines (CCXXCC, where X is any non-cysteine amino acid) that reacts with biarsenical dyes such as FlAsH (green, based on fluorescein) or ReAsH (red, based on resorufin).
SNAP-tag: In 2003, Kai Johnsson’s lab developed a self-labeling fusion tag based on the human DNA repair protein AGT that gets covalently linked to derivatives of its substrate O6-benzylguanine (BG) (Keppler et al., 2003). This engineered hAGT later became known as SNAP-tag. There is also a fast-reacting version of SNAP-tag called SNAPf.
eDHFR: The Virginia Cornish and Michael Sheetz labs demonstrated in 2005 that proteins fused with E. coli dihydrofolate reductase (eDHFR) could be labeled with fluorescent derivatives of its substrate trimpethoprim (TMP-tag) (Miller et al., 2005). Although this tight interaction is not covalent, later work showed that introducing a Leu28Cys mutation in eDHFR does lead to covalent binding of the TMP-tag ligand (Gallagher et al., 2009).
HaloTag: Scientists at Promega developed HaloTag in 2008 (Los et al., 2008), based on a bacterial haloalkane dehalogenase that binds a synthetic chloralkane (CA) ligand. The current standard version known as HaloTag is actually HaloTag7, and recent specialized variants run up to HaloTag11. HaloTag is now the most popular self-labeling tag in Addgene’s collection.
CLIP-tag: Also in 2008, Kai Johnsson’s lab reported another AGT-based tag named CLIP-tag, which reacts with O2-benzylcytosine(BC) ligands (Gautier et al., 2008). Although based on the same original enzyme, SNAP-tag and CLIP-tag show good specificity for their own ligands with minimal cross-reactivity.
Other strategies include using engineered ligases to attach a ligand to a peptide tag on your protein of interest or using non-natural amino acids to directly incorporate a fluorophore in your protein or to incorporate an alkene to be labeled through click chemistry. Although these multi-component systems can be complicated to implement, they can be highly specific with a small and minimally-disruptive genetic tag on the protein of interest.
Table 1: Summary of popular self-labeling tag systems
|
Tag |
Size (amino acids) |
Ligand |
Commercial ligands available |
Ease of use |
|
15 |
Biarsenical dyes (FlAsH, ReAsH) |
Few |
Easy |
|
|
182 |
Benzylguanine (BG) |
Many |
Easy |
|
|
159 |
Trimpethoprim (TMP-tag) |
Few |
Easy |
|
|
295 |
Chloralkane (CA) |
Many |
Easy |
|
|
182 |
Benzylcytosine (BC) |
Some |
Easy |
|
|
Small, varies |
Various |
Few |
Complicated |
|
|
1 |
Various |
Many |
Complicated |
Applications
Organic dyes are typically both brighter and more photostable than fluorescent proteins (often up to 10-fold more). So, labeling a HaloTag or SNAP-tag construct can help you better detect dim structures or low levels of expression with the same imaging workflows you already use. Or you can really capitalize on the dye’s performance and use super-resolution or single-molecule techniques.
 |
|
Figure 2: Two-color super-resolution imaging of ER and mitochondria reveals dynamic organelle interactions. a) Labeling scheme. b) COS-7 cell ER labeled with Halo-Sec61β with SiR-CA (magenta) and outer mitochondrial membrane labeled with SNAP-OMP25 with Atto590-BG (green), imaged with stimulated emission depletion (STED) nanoscopy. Scale bar, 2 μm. Inset shows comparison with standard confocal microscopy. Image adapted from (Bottanelli et al., 2016) under CC-BY license. |
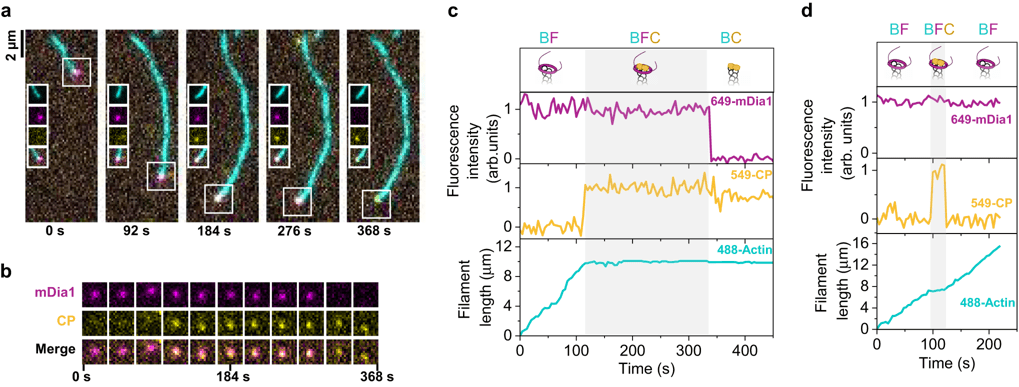
Combining orthogonal self-labeling tags on different proteins of interest (or one self-labeling tag and a fluorescent protein) enables applications like multi-color imaging (Figure 2). If you’re working with purified proteins in vitro, you can even use the same tag for multiple proteins but label each separately before reconstituting, choosing your colors depending on the needs of your experiment (Figure 3).
And using self-labeling tags makes updating your labels easy! Instead of taking the time to clone a new construct for every new-and-improved fluorescent protein you want to try, just swap in a new fluorescent ligand and see the results right away. Or perform pulse-chase experiments by sequentially adding different ligands to the same sample to label sub-populations of your protein of interest. For example, the Cohen lab used HaloTag for their "ticker tape" method for intracellular recordings, switching between different colored ligands at different time points.
 |
|
Figure 3: Single-molecule tracking of actin filament assembly-regulating proteins in vitro reveals complex events. a) Micrographs of a fluorescently-labeled actin filament (488-Actin, cyan), with formin (649-mDia1, magenta) and capping protein (549-CP, yellow) at the tip. 649-mDia1 and 549-CP are both SNAP-tag fusions, purified and labeled separately with different dyes. b) mDia1 and CP appearance at the filament tip from (a). c) Intensity traces at the tip of the filament from (a-b), showing the appearance and disappearance of 649-mDia1 or 549-CP (upper, middle) and the effects on actin filament length (lower). d) Intensity and filament length traces for a different event where CP appears briefly then leaves. Images reused from (Ulrichs et al., 2023) under CC-BY license. |
But these tags aren’t limited to fluorescent ligands! Self-labeling tags can also be used for surface immobilization, purification, or other uses of your protein, such as with substrate-linked PEG, biotin, or magnetic beads. In one innovative example, the Zhang lab used SNAP-tag to decorate synthetic viral particles with BG-linked antibodies to drive cell type-specific delivery of a viral vector (Strebinger et al., 2023).
All this flexibility comes at a price, though. Purchasing lots and lots of organic dyes or specialized ligands is more expensive than letting the cells make a fluorescent protein for you. And some ligands just aren’t going to work in certain applications — the most common limitation is that certain probes are not very good at crossing the cell membrane. So, you should investigate your options and weigh the potential benefits and costs before investing in one approach or the other.
Finding the right tag for you
Protein-based tags typically provide better labeling specificity than short peptide tags like tetracysteine (Jing & Cornish, 2011). HaloTag and SNAP-tag are the most popular self-labeling enzymes in Addgene’s collection, so you may be able to get a plasmid ready-to-use without any cloning at all! As mentioned above, several variants of these tags have been developed over the years; improving the labeling kinetics or the fluorophore’s microenvironment to enhance brightness, stability, or other properties is an active area of research.
If size is a concern for your application, SNAP-tag, CLIP-tag (both 182 aa), and eDHFR (159 aa) are smaller than HaloTag (295 aa) – for comparison, common fluorescent proteins are typically 230–250 aa. Smaller size isn’t a guarantee that a tag won’t disrupt the function of the target protein, so it’s always good to check that your fusion protein is functional. In some cases, a small peptide tag like tetracysteine might be the only option that doesn’t disrupt function.
If the idea of synthesizing your own ligands brings up bad memories of undergraduate organic chemistry lab, don’t worry – many SNAP-tag and HaloTag ligands are available from suppliers like New England Biolabs and Promega (though if you are comfortable with it or can find a colleague to help you, synthesizing your own ligands can open up more possibilities and save money). There aren’t as many commercial options for CLIP-tag and very few TMP-tag or tetracysteine labeling dyes.
If reaction speed is a priority, SNAP-Tag or HaloTag have been shown to perform differently for different ligands (Wilhelm et al., 2021). While HaloTag reacts faster for some dyes, others, especially some nonfluorescent ligands, react more efficiently with SNAP-tag. If you want to use far-red rhodamine dyes for super-resolution imaging, HaloTag gives better performance than SNAP-tag (Erdmann et al., 2019).
Check out our recent Illuminating Choices blog post for more discussion about choosing a dye or ligand to use for your experiment, and explore Addgene's collection to find self-labeling fusion proteins or cloning backbones to study your gene of interest with these tools.
Conclusion
Researchers and vendors continue to develop new ligands for these systems, enhancing their performance and ease of use. With so many possibilities from just one or a few constructs, it’s no wonder that self-labeling protein tags are increasingly popular. Embrace the flexibility and switch things up for your next experiments!
References and resources
References
Bottanelli, F., Kromann, E. B., Allgeyer, E. S., Erdmann, R. S., Wood Baguley, S., Sirinakis, G., Schepartz, A., Baddeley, D., Toomre, D. K., Rothman, J. E., & Bewersdorf, J. (2016). Two-colour live-cell nanoscale imaging of intracellular targets. Nature Communications, 7(1), Article 1. https://doi.org/10.1038/ncomms10778
Erdmann, R. S., Baguley, S. W., Richens, J. H., Wissner, R. F., Xi, Z., Allgeyer, E. S., Zhong, S., Thompson, A. D., Lowe, N., Butler, R., Bewersdorf, J., Rothman, J. E., Johnston, D. S., Schepartz, A., & Toomre, D. (2019). Labeling Strategies Matter for Super-Resolution Microscopy: A Comparison between HaloTags and SNAP-tags. Cell Chemical Biology, 26(4), 584-592.e6. https://doi.org/10.1016/j.chembiol.2019.01.003
Gallagher, S. S., Sable, J. E., Sheetz, M. P., & Cornish, V. W. (2009). An In Vivo Covalent TMP-Tag Based on Proximity-Induced Reactivity. ACS Chemical Biology, 4(7), 547–556. https://doi.org/10.1021/cb900062k
Gautier, A., Juillerat, A., Heinis, C., Corrêa, I. R., Kindermann, M., Beaufils, F., & Johnsson, K. (2008). An Engineered Protein Tag for Multiprotein Labeling in Living Cells. Chemistry & Biology, 15(2), 128–136. https://doi.org/10.1016/j.chembiol.2008.01.007
Griffin, B. A., Adams, S. R., & Tsien, R. Y. (1998). Specific Covalent Labeling of Recombinant Protein Molecules Inside Live Cells. Science, 281(5374), 269–272. https://doi.org/10.1126/science.281.5374.269
Jing, C., & Cornish, V. W. (2011). Chemical Tags for Labeling Proteins Inside Living Cells. Accounts of Chemical Research, 44(9), 784–792. https://doi.org/10.1021/ar200099f
Keppler, A., Gendreizig, S., Gronemeyer, T., Pick, H., Vogel, H., & Johnsson, K. (2003). A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nature Biotechnology, 21(1), 1. https://doi.org/10.1038/nbt765
Lavis, L. (2016, September 8). Better Dyeing Through Chemistry & Small Molecule Fluorophores. Addgene Blog. https://blog.addgene.org/better-dyeing-through-chemistry-small-molecule-fluorophores
Los, G. V., Encell, L. P., McDougall, M. G., Hartzell, D. D., Karassina, N., Zimprich, C., Wood, M. G., Learish, R., Ohana, R. F., Urh, M., Simpson, D., Mendez, J., Zimmerman, K., Otto, P., Vidugiris, G., Zhu, J., Darzins, A., Klaubert, D. H., Bulleit, R. F., & Wood, K. V. (2008). HaloTag: A Novel Protein Labeling Technology for Cell Imaging and Protein Analysis. ACS Chemical Biology, 3(6), 373–382. https://doi.org/10.1021/cb800025k
Miller, L. W., Cai, Y., Sheetz, M. P., & Cornish, V. W. (2005). In vivo protein labeling with trimethoprim conjugates: A flexible chemical tag. Nature Methods, 2(4), Article 4. https://doi.org/10.1038/nmeth749
Strebinger, D., Frangieh, C. J., Friedrich, M. J., Faure, G., Macrae, R. K., & Zhang, F. (2023). Cell type-specific delivery by modular envelope design. Nature Communications, 14(1), Article 1. https://doi.org/10.1038/s41467-023-40788-8
Ulrichs, H., Gaska, I., & Shekhar, S. (2023). Multicomponent regulation of actin barbed end assembly by twinfilin, formin and capping protein. Nature Communications, 14(1), Article 1. https://doi.org/10.1038/s41467-023-39655-3
Wilhelm, J., Kühn, S., Tarnawski, M., Gotthard, G., Tünnermann, J., Tänzer, T., Karpenko, J., Mertes, N., Xue, L., Uhrig, U., Reinstein, J., Hiblot, J., & Johnsson, K. (2021). Kinetic and Structural Characterization of the Self-Labeling Protein Tags HaloTag7, SNAP-tag, and CLIP-tag. Biochemistry, 60(33), 2560–2575. https://doi.org/10.1021/acs.biochem.1c00258







Leave a Comment