If you study proteins, you’re probably quite interested in the canonical 20 amino acids. But in your quest to learn more about your protein of interest, you may find the available amino acids a bit…limiting. It may be time, then, to look towards genetic code expansion (GCE): techniques in which non-canonical amino acids (ncAAs, also sometimes called unnatural amino acids) are placed in a protein site-specifically to alter its function, mimic a biologically relevant modification, or add in fluorescent or chemically reactive side chains, all of which can help determine structure and function relationships or, in more practical applications, allow a protein to be more effectively used in a technology or as a therapeutic.
A (very) brief history
GCE came onto the synthetic biology scene in 1989 through a seminal paper from Peter Schultz’s lab describing a technique that allowed for intentional incorporation of an unnatural amino acid in translation (Noren et al., 1989). In the late nineties, this was developed into an E. coli system, with the noncanonical amino acid in the media and the tRNAs and synthetase introduced to the bacteria using plasmids. Rapid expansion occurred, with hundreds of new ncAAs and techniques being developed. In the mid-aughts, the first yeast GCE paper was published, followed soon by GCE done in mammalian cells (Shandell et al., 2021).
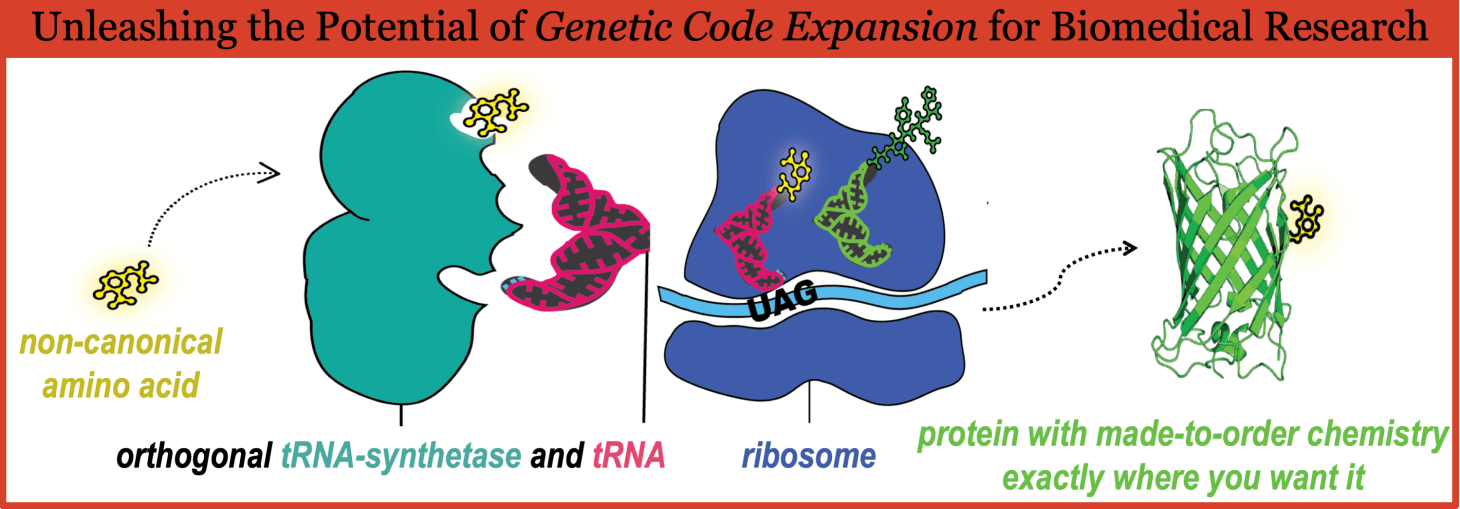
How it works
If you’re interested in using GCE, here are the basic requirements of use.
First, and most obviously, you need a non-canonical amino acid, which is usually added to the media for uptake by the cells.
Second, you’ll need tRNAs and tRNA synthetases specific to the ncAA and orthogonal to (i.e. non-interacting with) the cells’ own natural translation machinery; these are often incorporated into the cells via plasmid.
Third, a second plasmid will be needed to introduce the gene of interest containing the chosen codon that will result in the incorporation of the non-canonical amino acid at the site of interest (figure 1). While the codon used is often one of the three stop codons – most commonly the UAG codon – in some systems, rare codons and four-amino acid frameshift codons are also used.
Note that some systems have the tRNA, tRNA synthetase, and/or a biosynthetic pathway for the ncAA integrated into the cell’s genome, instead of incorporation via plasmid or media.
Theoretically, if you put all these together, the cell will produce a protein with a site specifically incorporated ncAA, ready for you do to any number of experiments with.
The catch
The rapid expansion of GCE led to the creation of many new tools and potential applications. But this focus on development, along with the inherent trickiness of using and optimizing GCE, led to a large toolbox filled with systems and ncAAs that were, in practice, mostly accessible to experts. That many GCE technologies have idiosyncrasies presents a significant barrier for those wishing to use it as a tool.
In short, while the technical skills needed to perform GCE are essentially those needed to make site-directed mutagenesis, many researchers new to GCE aren’t able to successfully complete their experiments and many others are dubious enough that they don’t give it a try.
Optimization
Why is GCE so complicated? Well, while the techniques are straightforward, the systems – the cells growing and producing the proteins – are not, and many of the existing GCE tools are not very robust.
When using GCE, each component of the experiment system (including media, cell type, and growth temperature) may need to be individually optimized for each of the three factors: the ncAA, the tRNA, and the tRNA synthetases. And, naturally, rarely are the optimization parameters the same for all three components, meaning that the final protocol may involve a sensitive balance of trade-offs. This balance can be extremely tricky to find: each of the factors can negatively impact both translation and fidelity of translation, leading to system-wide effects. And on top of that you need to worry about getting high enough expression of your protein of interest.
If that wasn’t enough, in this development-focused field, it’s not uncommon to find protocols that require reagents which aren’t broadly or easily available, so you may find yourself wanting to use a protocol that you’ll have to rework to include reagents accessible to your lab.
The solution
GCE provides almost endless possibilities for researchers, from selective protein ligation through introducing ncAAs with protein modifications of interest or any of a huge variety of ncAAs to study the relationship between structure and function…if you can get your protocol to work. Or, to quote Ryan Mehl, PhD, GCE has “10x the impact of PCR. But it’s 100x more complicated.”
And Ryan Mehl would know. He’s the director of GCE4All, an NIH-funded center located at Oregon State University focused on turning GCE tools developed in the literature into robust and universal techniques that researchers can easily implement in their own labs. They provide resources, knowledge, and most importantly, validated protocols (using readily available reagents!) for the use of GCE. In short, they’re working to make GCE available for all biomedical researchers.
If you’re interested in using GCE in your experiments, GCE4All is a great place to start. They host monthly webinars, workshops, and a bulletin board on their website specifically so people can talk about GCE.
In fact, their first protocol is available now. It is for recombinant expression of phosphoserine-including proteins in E. coli (Zhu et al., 2022). It uses the TAG codon for the phosphoserine and, because it uses ∆serB strains, does not require exogenously added phosphoserine.
A community resource
Like Addgene, GCE4All is a community resource which depends on engagement from the scientific community to fulfill their mission. While they’ve shortlisted several protocols, including their phosphoserine ncAA protocol, to optimize and share, they’re soliciting feedback from scientists on which protocols would be broadly useful for biomedical researchers. One way of doing that is by using Addgene’s Blue Flame award to identify highly requested GCE plasmids. But they’d also love scientist interested in, or currently using, GCE to reach out to them and let them know directly what protocols would be useful.
Think GCE could be useful in your research? We definitely recommend checking out GCE4All’s website and resources, and then checking out all of the plasmids and bacterial strains (available at Addgene for GCE work. Plus, keep an eye on our blog – we’ll be posting more posts on the various protocols and helping to spread the word as new protocols come out!
References
Noren, C. J., Anthony-Cahill, S. J., Griffith, M. C., & Schultz, P. G. (1989). A General Method for Site-specific Incorporation of Unnatural Amino Acids into Proteins. Science, 244(4901), 182–188. https://doi.org/10.1126/science.2649980
Shandell, M. A., Tan, Z., & Cornish, V. W. (2021). Genetic Code Expansion: A Brief History and Perspective. Biochemistry, 60(46), 3455–3469. https://doi.org/10.1021/acs.biochem.1c00286
Zhu, P., Mehl, R., & Cooley, R. (2022). Site-specific Incorporation of Phosphoserine into Recombinant Proteins in Escherichia coli. 12(21). https://doi.org/10.21769/BioProtoc.4541
Topics: Synthetic Biology, Other Plasmid Tools, Plasmids







Leave a Comment