 Every few months we highlight a subset of the new plasmids in the repository through our hot plasmids articles. These articles provide brief summaries of recent plasmid deposits and we hope they'll make it easier for you to find and use the plasmids you need. If you'd ever like to write about a recent plasmid deposit please sign up here.
Every few months we highlight a subset of the new plasmids in the repository through our hot plasmids articles. These articles provide brief summaries of recent plasmid deposits and we hope they'll make it easier for you to find and use the plasmids you need. If you'd ever like to write about a recent plasmid deposit please sign up here.
Listen to the Hot Plasmids episode!
Expanding the optogenetics toolbox with CRY2clust
Article contributed by Brook Pyhtila
![]() Listen to the CRY2clust podcast segment
Listen to the CRY2clust podcast segment
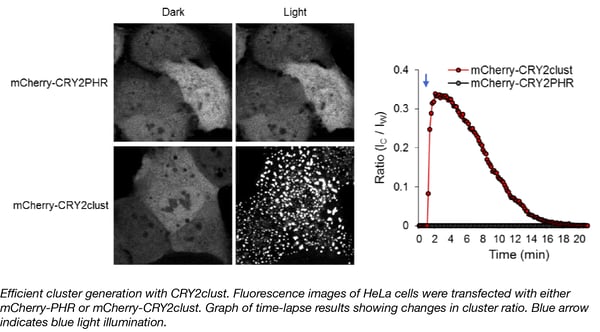
The Won Do Heo lab at the Korea Advanced Institute of Science and Technology (KAIST) and Institute for Basic Science (IBS) has developed another useful optogenetic tool that enables robust and efficient oligomerization of target proteins in response to blue light. This tool, CRY2clust, was created by adding a 9-residue peptide to the C-terminus of human codon-optimized cryptochrome 2 (CRY2) from Arabidopsis thaliana. When exposed to blue light, CRY2 undergoes a conformational change that permits it to bind to the CIB1 (cryptochrome-interacting basic-helix-loop-helix) protein. After fusing the CRY2 and CIB1 domains to separate proteins of interest, a researcher can cause them to interact by stimulation with blue light. Similarly, light can also be used to control homo-oligomerization of a protein fused to CRY2, although some studies have shown that this clustering only happens under certain conditions.
With its additional 9 amino acid peptide, the CRY2clust domain enables fast and reversible oligomerization when fused to a protein of interest. Fusion protein oligomerization happens within seconds after a pulse of blue light, and is reversible upon termination of light stimulation. CRY2clust enables faster assembly and disassembly dynamics, higher sensitivity to light, and better distribution of clusters without accumulation in subcellular compartments such as nuclear speckles. Furthermore, CRY2clust, when integrated into a Ca2+ -modulating optogenetic system (OptoSTIM1), caused two-fold faster changes in intracellular Ca2+ levels than the original version containing CRY2 without the extra peptide. CRY2clust should prove a useful tool to further probe cellular function.
The CRY2clust Plasmid can be found here.
Park et al. Nat Commun. 2017. PubMed PMID: 28646204.
soCoChR: A novel opsin that enables single-cell optogenetics
Article contributed by Meghan Rego
![]() Listen to the soCoChR podcast segment
Listen to the soCoChR podcast segment
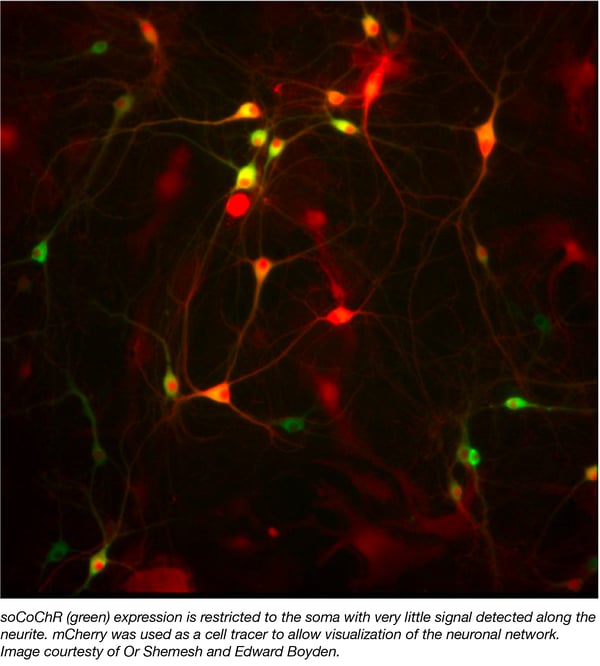
Optogenetic stimulation is a powerful tool used by neuroscientists to interrogate neuronal networks. A major drawback of this method is the stray excitation of neighboring neurons in opsin dense regions. To overcome this issue, Shemesh et. al. created a soma-targeted derivative of the opsin CoChR, termied soCoChR, by fusing a fragment of the kainate receptor subunit to the C-terminus of the protein. Unlike the diffuse neuronal expression observed with wild type CoChR, soCoChR localizes primarily to the cell body with very little signal detected along the neurite (Figure 1.) Moreover, the group was able to demonstrate that while illumination of neighboring cells evokes action potentials (APs) in patched cells expressing wild type CoChR, these APs are absent in soCoChR-expressing cells. Finally, using synaptic receptor blockers to distinguish real and artifactual postsynaptic currents (PSCs) this work shows that the artifactual PSCs commonly observed with direct stimulation are largely absent in the soCoChR-expressing system. Taken together, Shemesh et. al. describe a novel system for the precise mapping of neuronal networks at the single-cell level.
Various soCoChR plasmids described in this work including the soCoChR-GFP plasmid as well as several derivatives encoding alternative promoters and/or fluorescent tags are available at Addgene.
- Shemesh OA, et. al. Nature Neuroscience. 2017. PubMed PMID: 29184208.
Synthetic decaffeination operon
Article contributed by Angela Kaczmarczyk
![]() Listen to the synthetic decaffeination operon podcast segment
Listen to the synthetic decaffeination operon podcast segment
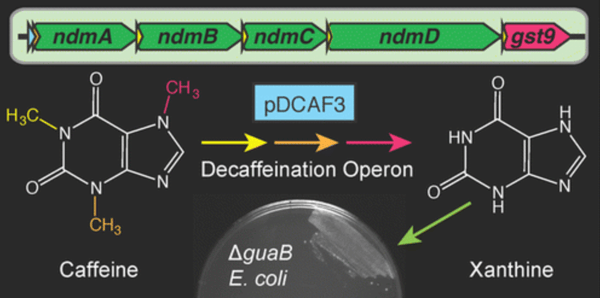
With the help of engineered microbes, used coffee grounds could be economically repurposed as starting material for the bioproduction of therapeutic methylxanthine drugs like theophylline. To facilitate future work with this goal, the Barrick lab has deposited the pDCAF3-IS5 plasmid containing a synthetic operon that enables the degradation of caffeine (1,3,7-trimethylxanthine) and its conversion into xanthine. The majority of this operon originates from Pseudomonas putida CBB5 and has been tailored for expression in E. coli. This operon is under the transcriptional control of a strong constitutive promoter (BBa_J23100) and expresses N-demethylases and other required enzymes, including a glutathione S-transferase homolog from a Janthinobacterium sp. Moreover, a caffeine “addicted” E. coli strain expressing this operon can serve as a living biosensor for measuring caffeine content in samples.
The Barrick lab and the UT Austin undergraduate iGEM team originally used a guaB knockout E. coli strain to assay for functional decaffeination in cells. The ΔguaB E. coli are missing an essential enzyme required for conversion of xanthine into guanine, and without xanthine supplied in the media, these bacteria are unable to divide. However, if fueled with caffeine and equipped with the pDCAF3 operon, the growth of this knockout strain can be rescued by the xanthine produced via complete demethylation of caffeine. For example, the Barrick lab was able to grow this strain in minimal media supplemented with various caffeinated beverages, but not with caffeine-free drinks. Finally, they demonstrated that the caffeine concentrations in several beverages could be estimated by measuring growth of the ΔguaB pDCAF3 strain.
Find the decaffeination plasmid here.
- Quandt et al. ACS Synth Biol. 2013. PubMed PMID: 23654268.
FLINC biosensors for live-cell imaging of biochemical activity
Article contributed by Angela Holmes
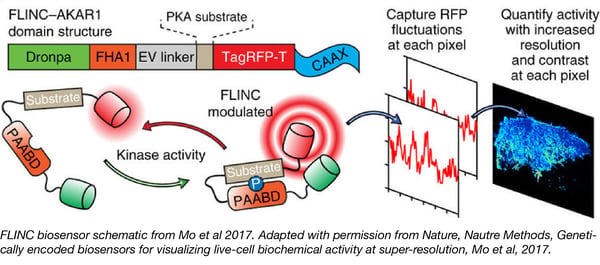
Jin Zhang’s lab is developing new super-resolution techniques for live-cell imaging of the dynamic changes in biochemical activity within the cell. Their Fluorescence fLuctuation INcrease by Contact (FLINC)-based biosensors measure fluctuations of TagRFP-T fluorescence intensity due to interactions with photoactivatable fluorescent protein Dronpa.
The authors tracked protein kinase A (PKA) activity at the plasma membrane in living cells using a construct in which TagRFP-T and Dronpa flanked a PKA substrate fused to the phosphopeptide binding domain FHA. The FHA1 domain binds to phosphorylated PKA substrate, inducing a conformational change that placesTagRFP-T and Dronpa in close proximity. By monitoring changes in TagRFP-T fluorescence intensity at different time points, the authors tracked protein activity while also capturing information about spatial and temporal organization.
The design use to detect PKA activity can be generalized to measure phosphatase activity, second messenger activity, and protein-protein interactions. We’re excited to see how you adapt similar FLINC biosensors to work in your lab.
Find the FLINC biosensors here.
- Mo et al. Nat Methods. 2017. PubMed PMID: 28288122.
New fluorescent proteins for use as FLIM-FRET acceptors
![]() Listen to the FLIM-FRET acceptor podcast segment
Listen to the FLIM-FRET acceptor podcast segment
Article contributed by Christina Mork
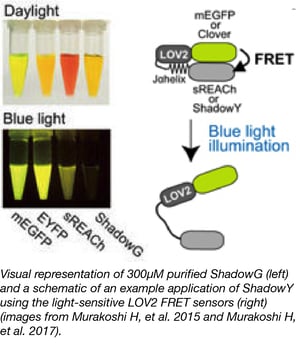
The Murakoshi lab has developed two new FLIM-FRET acceptors, ShadowG, a novel dark green fluorescent protein (GFP) with an absorption peak of 486 nm and ShadowY, a novel dark yellow fluorescent protein (YFP) with an excitation peak of 519 nm. These proteins have improved photophysical properties and can be used in quenching, FRET (Förster resonance energy transfer), and FLIM (fluorescence lifetime imaging microscopy) assays.
ShadowG is darker than other dark YFP mutants, it reduces the risk of spectral contamination artifacts, it has better folding and maturation kinetics than other dark fluorescent proteins, it has good signal stability allowing for minimized basal FRET variability, and is a good choice for FRET assays where a dark acceptor is desired due to its very low quantum efficiency.
ShadowY has a lower quantum yield and higher absorption than it’s predecessor, sREACH, allowing for reduced artifacts due to residual fluorescence. ShadowY was additionally shown to have a 7-fold lower quantum yield and a 1.2-fold greater extinction coefficient when compared to sREACH.
Find the FLIM-FRET acceptors here.
See Addgene's FRET reference page for many other common FRET vectors.
-
Murakoshi H, et al. Sci Rep. 2015. Pubmed PMID: 26469148.
-
Murakoshi H, et al. Sci Rep. 2017. Pubmed PMID: 28754922.
Topics: Other Plasmid Tools, Plasmids





Leave a Comment