Every few months we highlight a subset of the new plasmids and viral preps in the repository through our hot plasmids articles. These articles provide brief summaries of recent plasmid deposits and we hope they'll make it easier for you to find and use the plasmids you need.
Here's what you'll find in this article:
 An improved cell-free system to examine biosynthetic pathways
An improved cell-free system to examine biosynthetic pathways- A new tool for enriching ABE-targeted cells
- The CRISPR Corner
- New ready-to-use viral preps
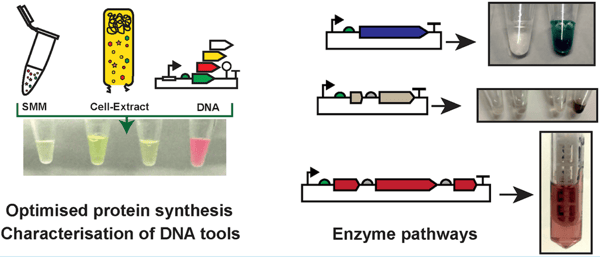
Improved cell-free transcription/translation system to examine biosynthetic pathways
Recently, cell-free transcription/translation systems (also referred to as TX-TL systems) have been gaining popularity as a synthetic biology tool. These systems allow researchers to understand bacterial gene clusters that produce useful natural materials, such as antibiotics. Simon Moore and Paul Freemont’s lab previously developed such a system in Streptomyces, a prokaryotic genus capable of producing a variety of natural products. Recently, this group optimized their initial system by using a stronger promoter and different biochemical secondary energy sources to increase protein yields from 36 μg/mL to 266 μg/mL. The reaction only required three components: a master mix, the DNA of interest, and cell extract.
 |
|
Graphical Abstract: Schematic representation of cell-free system, capable of producing fluorescent proteins and enzymes. Image from Moore et al., 2021. |
This TX-TL system is capable of robust expression of high G+C content genes, as validated through monitoring the expression of fluorescent proteins and oxytetracycline enzymes. Next, this system was used to recreate natural biosynthetic pathways. Biosynthetic pathways containing multiple enzymes could be successfully reconstituted in this cell-free system. In summary, this cell free system allows for a relatively simple, controlled “one-pot” reaction capable of reproducing useful natural materials.
Moore et al., ACS Synthetic Biology 202. https://doi.org/10.1021/acssynbio.0c00581
New tool for enrichment of ABE-targeted cells
By Ellie Adams
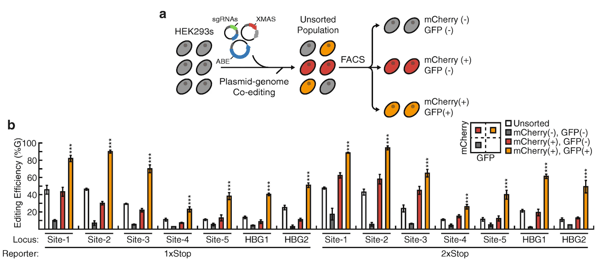
Single base modifications through ABE or CBE's have the potential ability to modify up to 60% of the disease-causing point mutations. They allow single-nucleotide modifications to edit DNA without requiring a double-stranded break. The Xiao Wang Lab has recently deposited a set of plasmids that helps identify base edited cell populations without end-point sequencing assays. These plasmids use “Cas9-mediated adenosine transient reporter for editing enrichment”, or XMAS-TREE, to identify base edited cell populations without end-point sequencing assays.
This tool was designed by putting together the coding sequence of a mCherry fluorescent protein followed by a stop codon and the coding sequence for a green fluorescent protein (GFP). The success of the A-to-G conversion could be seen through the success of the change in expression of GFP, for the stop codon should have changed from “TGA” to “TGG.” After identification of ABE activity, XMAS-TREE can also be used to purify these modifications and work with them further. The lab demonstrated that XMAS-TREE can be multiplexed and can be used in human pluripotent stem cells, a cell type that has been difficult to modify in the past.
Brookhouser, N., et al. BMC Biol 18, 193 (2020). https://doi.org/10.1186/s12915-020-00929-7
The CRISPR corner 
New CRISPR plasmids are always being added to the repository. To find all of the CRISPR plasmids available from Addgene, head over to our CRISPR Plasmids and Resources page. Here are some highlights from the past couple months including several pooled libraries:
- Cas13X and Cas13Y, two compact Cas13 proteins found in the metagenome datasets from hypersaline samples, were engineered and adapted for RNAi experiments for mammalian cell lines.
- The Turner Lab Human messenger-RBP sgRNA Library targets human mRNA binding proteins with 10 guide RNAs per gene. The lab modified the lentiviral backbone to encode the CD90.1 selection marker, Thy1.1, in addition to puromycin resistance.
- The MYC-CRISPR library targets E-boxes genome-wide. It was designed based on MYC-ChIP-seq data from several MYC-dependent cancer cell lines. It can be applied for identification of essential MYC binding sites in a wide range of human cells.
- A new pooled library from Junjie Chen's lab targets human genes known or suspected to be involved in DNA damage response and DNA repair.
- A new CRISPR pooled library targeting S. pneumoniae D39V from Jan-Willem Veening is now available with one sgRNA per operon or gene. The sgRNA pool can also target core operons in other pneumococcal strains.
- The BARBEKO sgRNA library uses CRISPR cytosine base editors to disrupt gene function by targeting start codons, splice acceptor and donor sites, and introducing premature stop codons.
New from the viral service
By Jason Nasse
We regularly add new viral aliquots from our plasmid collection to provide ready-to-use viral preps. Here are some of the new viral preps from recent months:
- Second generation GRAB dopamine sensors from Yulong Li’s lab are now available including:
- pAAV-hsyn-GRAB_DA2h in AAV9
- New GRAB-DA and 5HT sensors
- New AAV tools to selectively control parvalbumin interneurons from the Dimidschstein Lab
- More GCaMP8 ready-to-use AAV preps

Topics: Hot Plasmids, Plasmids







Leave a Comment