Part stable scaffold, part epitope binding region, all fused to a fluorescent protein. “Frankenbody,” like the infamous frankenstein, was constructed by grafting different parts together. By combining two optimized elements of an antibody probe, scientists can now more easily visualize single molecules and newly formed proteins.
Limitations with GFP in imaging
While GFP has been an immensely powerful tool to study proteins in vivo, it does have some limitations. GFP has long protein maturation times which limits its use for imaging a protein’s entire life-cycle. This makes it nearly impossible for scientists to use GFP to image nascent proteins. In addition GFP tags are large, dim, and permanently attached to a protein, making them hard to amplify and image for long periods of time.
Are antibody based probes a solution?
To address these limitations, scientists have co-opted antibodies for tagging purposes. Different antibodies bind different protein sequences called epitopes and scientists can use these to tag their proteins of interest. Antibody based probes, such as antigen-binding fragments (Fabs) (Porter 1959) and single-chain variable fragments (scFvs) (Inbar et al., 1972) are fused to mature fluorophores, and can bind and light up epitope tagged proteins in vivo. These probes however can be expensive to make, hard to deliver into living systems, and often do not fold properly once inside cells.
Developing the HA frankenbody
To combat the issues with these antibody based probes the Stasevich lab used an alternative strategy to develop a genetically encoded antibody based probe that detects the widely used HA epitope in diverse cellular environments. They called this probe the HA frankenbody (Zhao et al., 2019).
To develop HA frankenbody, the Stasevich lab tested several scFvs scaffolds previously proven to fold and function in living cells. To make these scaffolds specific to different targets, the scientists fused them to sequences called complementarity determining regions (CDRs, or loops) of an antibody that recognize and bind specific epitopes (Ewert et al., 2004). This scFv hybrid provides the probe structure and stability from the scaffold as well as specificity towards a HA epitope from the CDRs.
To visualize proteins of interest tagged with the HA epitope, the lab fused a fluorescent protein to the hybrid anti-HA scFvs. The addition of the fluorescent protein allows scientists to easily observe the tagged protein upon probe binding in living cells.
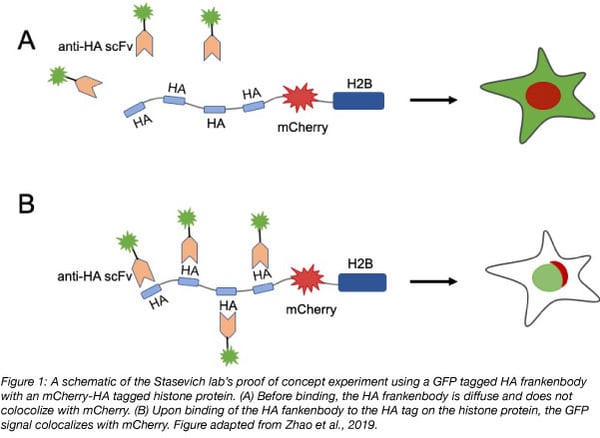
In their proof of concept experiment the Stasevich lab observed colocalization of GFP tagged HA frankenbody with an mCherry, 4xHA tagged histone protein (H2B) in U2OS cells. They then showed that the HA frankenbody probe could also bind a 1xHA tagged histone protein as well as a mitochondrial protein tagged with spaghetti monster HA (smHA) which contains a 10xHA tag. These experiments demonstrated that the HA frankenbody can be used to amplify fluorescent signals in in vivo imaging: the 10xHA tag increased the fluorescent signal to noise ratio five fold over the 1xHA tag.
 Lastly, the researchers found that the HA frankenbody is effective in sensitive cells such as living neurons, and can be fused to other fluorescent proteins and tags including mCherry, HaloTag and SNAP-tag for multi-color imaging.
Lastly, the researchers found that the HA frankenbody is effective in sensitive cells such as living neurons, and can be fused to other fluorescent proteins and tags including mCherry, HaloTag and SNAP-tag for multi-color imaging.
Find the HA frankenbody tool-kit at Addgene to get started with your experiments!
What else can you do with the HA frankenbody?
Another large advantage of the HA frankenbody is that it remains bound to HA epitopes for minutes in living cells, meaning that scientists can track the movement and localization of single HA-tagged molecules over time.
The Stasevich lab also proved that HA frankenbody can be effectively used to track single mRNA translation in U2OS cells. HA is small, only 9 amino acids, thus it is translated quickly by the ribosome. This feature provides HA frankenbody the ability to bind nascent HA tagged proteins.
Lastly, the HA frankenbody can be used for imaging in whole organisms. The researchers used the HA frankenbody to successfully track nuclei in a developing zebrafish embryo over an 80 minute time course.
By stitching together a stable scaffold and HA-binding regions, the Stasevich lab created a versatile tool that can be used to visualize a wide range of protein dynamics in vivo. In the future, this stitching technique could be applied to other epitopes and scaffolds to build a more robust set of frankenbodies.
References
Ewert, S., Honegger, A., & Pluckthun, A. (2004). Stability improvement of antibodies for extracellular and intracellular applications: CDR grafting to stable frameworks and structure-based framework engineering. Methods, 34(2), 184–199. PubMed PMID: 15312672.
Inbar, D., Hochman, J., & Givol, D. (1972). Localization of antibody-combining sites within the variable portions of heavy and light chains. Proceedings of the National Academy of Sciences of the United States of America, 69(9), 2659–2662. PubMed PMID:4560694. PubMed Central PMCID: PMC427010.
Porter, R. (1959). The hydrolysis of rabbit y-globulin and antibodies with crystalline papain. The Biochemical Journal, 73(1948), 119–126. PubMed PMID: 14434282. PubMed Central PMCID: PMC1197021.
Zhao, N., Kamijo, K., Fox, P. D., Oda, H., Morisaki, T., Sato, Y., … Stasevich, T. J. (2019). A genetically encoded probe for imaging nascent and mature HA-tagged proteins in vivo. Nature Communications, 10(1). PubMed PMID: 31270320. PubMed Central PMCID: PMC6610143.
Additional resources on the Addgene blog
- Learn about visualizing protein turnover
- Download the fluorescent protein eBook
- Browse our fluorescent protein blog posts
Resources on Addgene.org
Topics: Fluorescent Proteins, Localization with Fluorescent Proteins





Leave a Comment