To respond to environmental conditions, cells must be able to detect extracellular stimuli. One way they do so is through G protein-coupled receptors (GPCRs), receptors that play many roles in signaling processes including smell, taste, sight, inflammation, and neurotransmission. But with approximately 950 different human genes encoding GPCRs (Takeda S et al., 2002) and many downstream complexes involved, it can be difficult for scientists to understand how GPCR activation leads to downstream effects.
Bryan Roth’s lab created a collection of plasmids called TRUPATH to interrogate 14 of the 16 non-visual G protein signal transducers (collectively known as the transducerome) that can be activated downstream of GPCRs that are not involved in sight (Olsen RHJ et al., 2020). A transducer complex consists of a trimer of three subunits: alpha, beta, and gamma (Gα, Gβ, and Gγ), collectively known as a heterotrimeric G protein. This complex converts a signal received by a GPCR on the cell’s surface into action that can result in the initiation of a vast number of signaling cascades, inducing different cellular responses. One of the biggest advantages of such a tool is being able to identify specific transducer complexes that are activated in response to a ligand binding a GPCR of interest.
How does TRUPATH work?
The TRUPATH collection allows researchers to measure the physical association of combinations of different Gα, Gβ, and Gγ subunits. When inactive, these subunits are found in a trimeric complex bound to the intracellular surface of the GPCR. Binding of a ligand to the extracellular surface of the GPCR causes a conformational change in the GPCR, leading to the activation of the Gα subunit. The activated Gα exchanges bound GDP with GTP, resulting in the disassociation of the Gα subunit from the Gβγ dimer, activating downstream signaling pathways.
This dissociation can be measured using a technique known as Bioluminescence Resonance Energy Transfer (BRET). BRET is similar to Förster Resonance Energy Transfer (FRET), in which a donor fluorophore is used to excite an acceptor fluorophore. The difference for BRET is that the energy transfer is between a luminescent donor and a fluorescent acceptor. TRUPATH uses an updated version of BRET called BRET2.
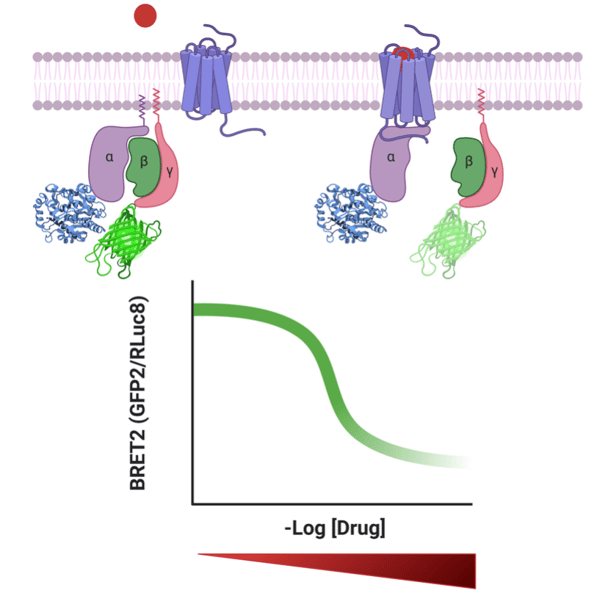
|
Figure 1: TRUPATH measures heterotrimeric G protein dissociation by bioluminescence resonance energy transfer 2, or BRET2. With binding of the drug, the alpha and beta subunits dissociate resulting in decreased BRET2 signal. With increased concentration of drug, the BRET2 signal decreases. |
The TRUPATH BRET2 biosensor system was developed by constructing 14 luciferase donor Rluc8-Gα chimeras, four acceptor GFP2-Gγ chimeras, and two untagged Gβ constructs. For the Gγ constructs, the GFP2 tag is fused to the N-terminus of the protein. For the Gα constructs, localization of the Rluc8 was more challenging. The Roth lab used structure-guided protein engineering and experimental refinement to determine the optimal localization of the Rluc8 insertion within the Gα. Despite exhaustive refinement, it was not possible to generate an optimal Rluc8-Gα construct for two of the 16 non-visual G protein transducers.
To use the TRUPATH system, cells are transfected with plasmids expressing the GPCR of interest, a Gα-Rluc8 plasmid, and the appropriate Gγ-GFP2 and Gβ constructs. Cells are then treated with the luminescent enzyme substrate coelenterazine, followed by the addition of the endogenous ligand or drug. In the inactive state, the luminescent chimeric protein Gα-Rluc8 and Gγ-GFP2 will be in close association, resulting in a high level of GFP2 emission. Following activation in response to the ligand, the complex will disassociate, resulting in a decreased GFP2 emission. The activation state can be measured by a BRET2 ratio, which is the ratio of GFP2 emission to RLuc8 emission (see figure).
What can TRUPATH be used for?
The Roth lab used the TRUPATH plasmids in a number of different experimental approaches. They first assessed the activation of different members of the transducerome for both well-studied and understudied GPCRs in response to different agonists. These studies showed the promiscuity of GPCRs for different G protein transducers and that the potency and efficacy is typically non-uniform across the transducerome for a specific receptor-ligand pair. They further used TRUPATH to analyze a panel of compounds across the transducerome on a single GPCR, showing the effectiveness of using TRUPATH as a screening technique. Lastly, the activity of both antagonists and inverse-agonists could be measured using the TRUPATH system.
Conclusions
Although the importance of GPCR pathways is well known and, as a result, a target of many pharmaceutical drugs, there is still only a partial understanding of how GPCRs signal through G protein signal transducers. The TRUPATH collection provides scientists with a toolkit to gain an understanding of the signaling through the transducerome from different GPCRs. This collection can be used to study endogenous ligands, the pathways they activate, as well as the activation by drugs to further understand their mechanism. These plasmids are available as a collection, facilitating screening across the transducerome, to gain a more complete understanding of a GPCR mediated response.

Acknowledgements
Special thanks to Jeff DiBerto, a graduate student in the Roth lab, for editing the blog post.
References
Olsen RHJ, DiBerto JF, English JG, Glaudin AM, Krumm BE, Slocum ST, Che T, Gavin AC, McCorvy JD, Roth BL, Strachan RT (2020) TRUPATH, an open-source biosensor platform for interrogating the GPCR transducerome. Nat Chem Biol 16:841–849 . https://doi.org/10.1038/s41589-020-0535-8
Takeda S, Kadowaki S, Haga T, Takaesu H, Mitaku S (2002) Identification of G protein-coupled receptor genes from the human genome sequence. FEBS Letters 520:97–101 . https://doi.org/10.1016/s0014-5793(02)02775-8
Additional resources on the Addgene blog
- Browse all of our fluorescent protein blog posts
- Find other fluorescent biosensors
Resources on Addgene.org





Leave a Comment