The late Michael Davidson and Roger Tsien played enormous roles in fluorescent protein development by openly sharing their resources with the research community. To honor their legacy, we launched the Michael Davidson and Roger Tsien Commemorative Travel Awards in 2017. These travel awards are intended to help early career scientists attend a conference of their choice to share their fluorescent protein based research.
This year, we put out another call for applications. We had an overwhelming response and were excited to see the many creative uses of fluorescent proteins. We’re happy to announce the three winners of this year’s Michael Davidson and Roger Tsien Commemorative Travel Awards: Melissa Chiasson, Elizabeth Unger, and Michal Bernach.
Melissa Chiasson: Quantifying the abundance of thousands of protein variants simultaneously
 With increasing sequencing power, scientists are able to quickly sequence an individual’s genome. But when they find genetic variants, what does that mean? Previous research shows that approximately 75% of all protein-coding pathogenic human variants are decreased in protein abundance, possibly due to misfolding. But measuring variants one at a time is time consuming and unscalable.
With increasing sequencing power, scientists are able to quickly sequence an individual’s genome. But when they find genetic variants, what does that mean? Previous research shows that approximately 75% of all protein-coding pathogenic human variants are decreased in protein abundance, possibly due to misfolding. But measuring variants one at a time is time consuming and unscalable.
Melissa Chiasson, a graduate student in Douglas Fowler's lab at the University of Washington sought to address the effects of genetic variants in a high throughput manner. She helped develop a method that measures the effects of sequence variants on abundance simultaneously called VAMP-seq, for Variant Abundance by Massively Parallel sequencing.
In VAMP-seq, the abundance of a protein variant library is monitored by fusing the variants to eGFP followed by an internal ribosomal entry site and mCherry. If a variant has wild-type levels of abundance, the eGFP will be stable and fluoresce. But if the variant is unstable, it will be degraded, leading to reduced eGFP fluorescence. Cells expressing the library are then sorted by fluorescence-activated cell sorting into four bins based on their eGFP:mCherry ratios. Genomic DNA from each bin are then extracted and sequenced.
Chiasson is currently using VAMP-seq to study vitamin K epoxide reductase (VKOR) variants. VKOR is an enzyme critical for activating clotting factors and is the target of the anticoagulant drug warfarin. In past research, several VKOR variants have been found that affect the body’s response to warfarin. “These novel methods will be instrumental in interpreting human variation at large scales and making personalized medicine a reality,” Chiasson explains.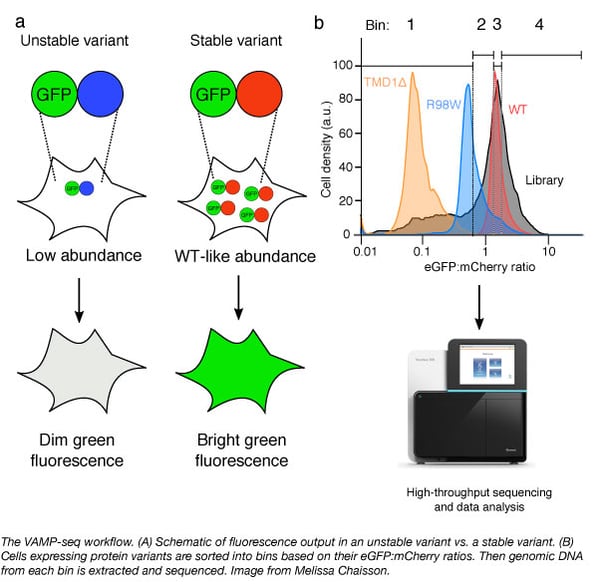
Elizabeth Unger: Developing a serotonin sensor
 The goal of Elizabeth Unger’s postdoctoral work in Lin Tian’s lab at UC Davis is to generate a genetically encoded fluorescent sensor for serotonin. According to Unger, “serotonin is a well-known but poorly studied neuromodulator, and evidence suggests that it is incredibly important for nearly every aspect of life.”
The goal of Elizabeth Unger’s postdoctoral work in Lin Tian’s lab at UC Davis is to generate a genetically encoded fluorescent sensor for serotonin. According to Unger, “serotonin is a well-known but poorly studied neuromodulator, and evidence suggests that it is incredibly important for nearly every aspect of life.”
Unger began with the a bacterial periplasmic binding protein, one similar to the protein which gave rise to the iGluSnFR sensor. However, there are no known bacterial proteins that bind serotonin. Unger redesigned the binding pocket of the periplasmic binding protein so that it would recognize serotonin. Using a combination of computer modeling, directed evolution, site-saturation mutagenesis, and medium-throughput screening, she increased the affinity of the sensor for serotonin by over four orders of magnitude.
Unger validated the serotonin sensor with proof-of-principle experiments that replicate current knowledge, but the sensor can also kinetically outperform current methods. Unger plans to use the sensor to answer the question of whether firing of serotonergic neurons (measured using a red calcium sensor) leads to the release of serotonin (measured using the newly developed green serotonin sensor).
In preliminary studies with the lab’s collaborators, they were able to use the serotonin sensor in sleep-related studies and in assays for measuring human serotonin transporter activity. Unger also plans to use the serotonin receptor to study how single nucleotide polymorphisms present in the human population can affect drug and dose response.
Selective serotonin reuptake inhibitors are frequently used to treat depression and anxiety, but these drugs are often ineffective or require several rounds of dose and drug adjustments. Unger ultimately hopes that medical professionals can genotype the patient and rapidly test the variants in vitro against different drugs and dosages. 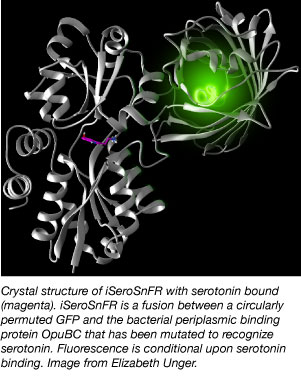
Michal Bernach: Developing an artificial leaf replica system to study plant-bacteria and bacteria-bacteria interactions
 Michal Bernach is a graduate student in Mitja Remus-Emsermann’s lab at the University of Canterbury where he studies how bacteria interact with each other and plant surfaces. His system of choice is the Arabidopsis thaliana - Pantoea agglomerans interaction.
Michal Bernach is a graduate student in Mitja Remus-Emsermann’s lab at the University of Canterbury where he studies how bacteria interact with each other and plant surfaces. His system of choice is the Arabidopsis thaliana - Pantoea agglomerans interaction.
But the shape and metabolism of leaves can be changed by light and temperature and leaf autofluorescence can interfere with microscopy. Thus, Bernach sought to develop an artificial leaf replica system. “This bottom-up approach will allow me to create a fully controllable and reproducible set up,” Bernach explains.
Developing an artificial leaf replica means creating a “leaf” with similar permeability of water and nutrients and colonization opportunities for bacteria. To compare the artificial leaf to A. thaliana leaf, Bernach plans to use a suite of fluorescent reporters. Bacteria containing unstable fluorescent proteins under the control of promoters responsive to water or nutrient levels can help him determine whether the artificial leaf behaves like the living leave in this aspect. To monitor bacterial colonization on the artificial leaf, Bernach will use the CUSPER bioreporter such that fluorescent proteins expression is induced prior to inoculation with IPTG. In the absence of IPTG after inoculation, no more fluorescent proteins are made. As the bacterial replicate, fluorescent proteins will be diluted out and the signal will diminish.
Once the system is validated, Bernach plans to use it study bacterial population dynamics with the TIMER growth rate bioreporter. During protein maturation, the bioreporter spontaneously changes from green to orange such that non-growing cells are orange, slow growing cells are orange-green, and fast growing cells are green. A second colonizer, labelled with a constitutively fluorescent reporter can also be used to track how one population responds to a second population.
Last, Bernach will simultaneously monitor cell cycle and growth rate using TIMER and an unstable fluorescent protein reporter under the control of promoters that control cell cycle gene expression.
Additional Resources on the Addgene Blog
- Visit the fluorescent protein topics page
- Get the basics on GFP
- Find microbiology blog posts
Resources on Addgene.org
- Find fluorescent protein biosensors
- Browse Plasmids from the Tsien lab
- Browse the Michael Davidson collection
Topics: Addgene News, Awards





Leave a Comment