 Fluorescent proteins have enabled scientists to pursue creative research avenues previously unavailable to them. With these fantastic tools it’s now easy to monitor protein expression, localization, and protein-protein interactions. Beyond these common applications, researchers are finding new ways to apply fluorescent proteins everyday.
Fluorescent proteins have enabled scientists to pursue creative research avenues previously unavailable to them. With these fantastic tools it’s now easy to monitor protein expression, localization, and protein-protein interactions. Beyond these common applications, researchers are finding new ways to apply fluorescent proteins everyday.
The late Michael Davidson and Roger Tsien played enormous roles in enabling researchers to utilize and develop these tools by making their own fluorescent protein technologies widely available to the research community. To honor their legacy, we recently launched the Michael Davidson and Roger Tsien Commemorative Travel Awards. These awards are intended to help scientists share their fluorescent protein research with the academic community by funding their travel to a conference of interest.
In this post, we’re proud to announce our first two Michael Davidson and Roger Tsien Commemorative Travel Awardees: Xin Zhou and Fatima Enam. There were many excellent applicants but Xin and Fatima stood out for their creativity in the use of fluorescent proteins and the potential for their work to enable future discoveries.
Read on to learn more about Xin and Fatima's work and keep checking back in with the Addgene blog for future award opportunities.
Xin Zhou: Fluorescent Light-inducible Proteins (FLiPs)
 Fluorescent proteins can be applied in a long list of creative ways. In her graduate work in Michael Lin’s lab at Stanford, Xin Zhou greatly expanded that list by using photoswitchable fluorescent proteins to create enzymes that can be turned on and off with light.
Fluorescent proteins can be applied in a long list of creative ways. In her graduate work in Michael Lin’s lab at Stanford, Xin Zhou greatly expanded that list by using photoswitchable fluorescent proteins to create enzymes that can be turned on and off with light.
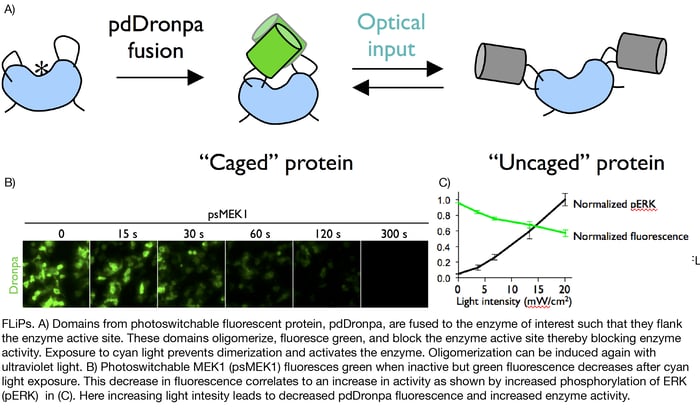
Xin hypothesized that she could make enzymes controllable by tethering them to Dronpa, a fluorescent protein known to oligomerize when exposed to ultraviolet light. To do so, she generated fusion proteins in which Dronpa domains flanked either side of an enzyme’s active site. In these fusions, ultraviolet light exposure caused oligomerization of the Dronpa domains thereby occluding the enzyme active site and shutting off the enzyme. Later exposure to cyan light stopped Dronpa oligomerization and turned the enzymes back on. Xin termed these photoswitchable enzymes Fluorescent Light-inducible Proteins, or FLiPs.
Xin later optimized these first generation FLiPs by mutating Dronpa to lower its aggregation tendency, increase its brightness, and decrease the amount of light required for dimerization. She additionally showed that these enhanced “pdDronpa” domains could be placed in amino acid loops on either side of an enzyme’s active site and still block enzyme function upon exposure to ultraviolet light. Previous iterations of FLiPs only used N- and C-terminal dronpa fusions.
With these various designs, Xin has been able to develop photoswitchable Intersectin, Cdc42, Raf1, Mek1, Mek2, Cdk5, and Cas9. Xin has demonstrated the utility of these FLiPs in a variety of research applications and particularly notes their ability to be precisely spatio-temporally controlled as a key advantage over other inducible enzyme technologies. She hopes that other researchers will be able to use similar designs to create a wide variety of enzymes and proteins with inducible activity and recently presented her work at the 2017 Winter qBio Meeting.
You can learn more about Xin’s work with photoswitchable components of the ERK signaling pathway in our recent blog post.
Fatima Enam: Biosensors for human milk oligosaccharides
 Fatima Enam is currently a graduate student in Thomas J. Mansell’s lab at Iowa State University where she is developing fluorescent protein-based biosensors for Human Milk Oligosaccharides (HMOs).
Fatima Enam is currently a graduate student in Thomas J. Mansell’s lab at Iowa State University where she is developing fluorescent protein-based biosensors for Human Milk Oligosaccharides (HMOs).
HMOs are thought to play an important role in nuturing the development of beneficial bacteria in the newborn gut. One of Fatima’s ultimate goals is to engineer bacteria for the efficient product of HMOs that could be used to supplement newborn diets and promote the formation of a healthy microbiome.
In order to achieve this goal, Fatima first needed an efficient and cost-effective way to measure the production of HMOs. Current HPLC-MS and fluorophore-based technologies are too time consuming in the first case and nonspecific in the second. Fatima therefore set out to design and produce two biosensors that could quickly and reliably detect HMOs.
Fatima’s first design is a whole cell biosensor with two components - an enzyme that breaks down the HMO of interest into its component simple sugars (including lactose), and a lac operon controlled GFP. When the enzyme breaks down the HMO of interest, the lactose released activates expression of GFP from the lac operon. GFP expression can then be used to determine the amount of HMO originally in the sample.
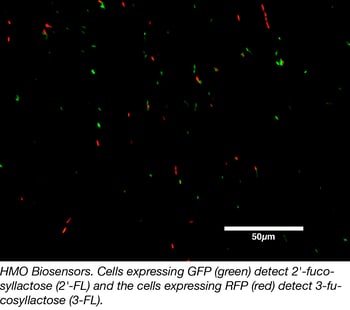 The second sensor is a fusion protein consisting of sfGFP fused to a His-tagged ABC transporter known to bind to the HMO of interest. Fatima specifically proposes using this sensor to detect HMOs found on cellular surfaces. After incubating the sensor with bacteria engineered to produce HMOs, sensor binding results in an increase in fluorescence of cells that actively produce the HMOs.
The second sensor is a fusion protein consisting of sfGFP fused to a His-tagged ABC transporter known to bind to the HMO of interest. Fatima specifically proposes using this sensor to detect HMOs found on cellular surfaces. After incubating the sensor with bacteria engineered to produce HMOs, sensor binding results in an increase in fluorescence of cells that actively produce the HMOs.
Fatima has demonstrated the successful production of both types of HMO biosensors and displayed her findings at the Spring 2017 ACS meeting. She hopes these biosensors and her future work with HMOs will enable researchers to engineer healthier microbiomes in people all over the world. Toward this end, she recently presented her work at her home town in Bangladesh where it was well received.
In addition to her research endeavors, Fatima hopes to practice her science communication skills throughout her training and wants to make her work understandable to people both inside and outside the scientific community. We’re excited to follow Fatima’s development as a researcher and science communicator.
Update (7/17/2018): Check out Fatima's recent publication on her HMO work in Cell Chemical Biology!
Additional Resources on the Addgene Blog
- Learn More about Photosactivatable Fluorescent Proteins
- Use Light to Control CRISPR Genome Editing
- Choose the Brightest Fluorescent Protein for Your Experiments
Resources on Addgene.org
- Find Fluorescent Protein Biosensors
- Browse Plasmids from the Tsien Lab
- Browse the Michael Davidson Collection
Topics: Addgene News, Awards






Leave a Comment