Signal transduction pathways are a lot like cell phone networks. Protein kinases deliver messages to the next members of their pathways, but where the kinases are located, their signal strength, and how long their signals last all impact transduction of the message. To study signaling pathways, scientists frequently use growth factors or serum to stimulate a pathway of interest, but there can be a lot of static since other signaling networks can also be non-specifically activated and, like the game of telephone, often the signal must be transmitted by intermediate messengers. Optical control of kinase activity can provide greater spatiotemporal resolution than pharmacological or genetic approaches, but only a few such methods exist and they only work for a subset of kinases.
The Problem:
There is no common method for creating optically controlled kinases.
The Solution:
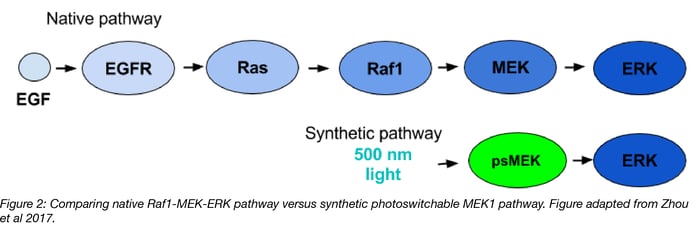
In Zhou et al, the Lin lab present a generic design for creating photoswitchable kinases and use MEK1 as a model kinase. In this system, light is used to turn “on” and “off” MEK1 activity by attaching two dimerizing photoswitchable Dronpa (dron= a ninja term for vanishing, and pa= photoactivation) domains on either side of the active site of a constitutively active MEK1. This photoswitchable MEK1 is called psMEK1. When exposed to 500 nm violet light, the Dronpa dimers interact and lock MEK1 in an “off” setting by blocking the active site. Dronpa also produces green fluorescence when exposed to 500 nm light. Under 400 nm cyan light, Dronpa dimers dissociate, Dronpa goes dark, and MEK1 is active or “on.” Dronpa’s color thus indicates MEK1’s current state (Green when off, dark when on).
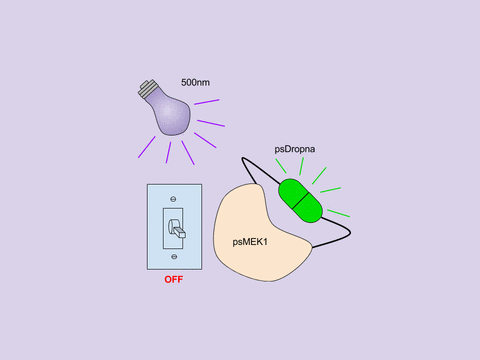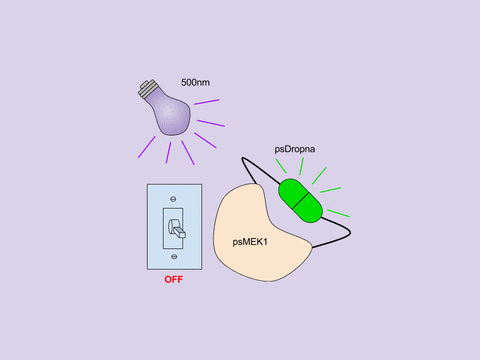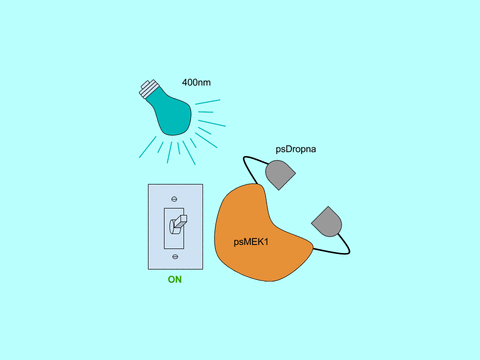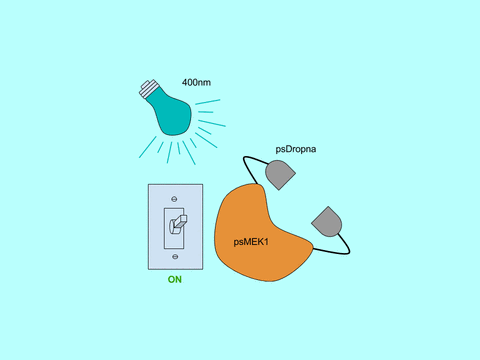
Figure 1: Photoswitchable MEK1 (psMEK1).
Is it possible to create other photoswitchable kinases?
The general design of psMEK1 also worked for creating three other photoswitchable kinases: psMEK2, psRaf1, and psCDK5. MEK2 is a serine/threonine kinase with a similar 3D structure to MEK1, but Raf1 and CDK5 have less similarity. For these three kinases, photoswitchable Dronpa domains were inserted at sites homologous to the locations they were attached to in MEK. The photoswitchable versions of MEK1, MEK2, Raf1, and CDK5 all behave similarly to their endogenous counterparts. Additionally, dimeric Dronpa is less prone to aggregation in cells compared to the tetrameric Dronpa it was derived from.
What are photoswitchable kinases good for?
There are a number of potential applications for photoswitchable kinases. Below are a few examples demonstrated in Zhou et al.
1. Testing kinase inhibitors
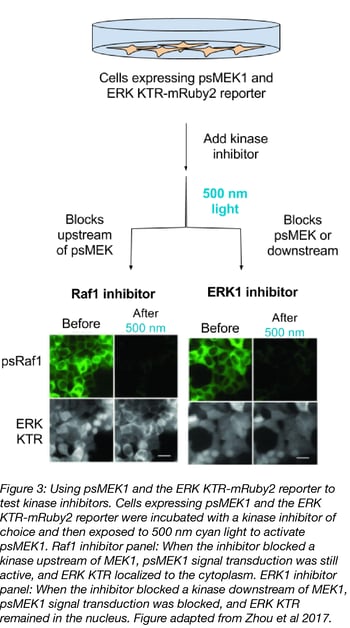
Zhou et al examined the ability of kinase inhibitors to alter Raf1-MEK-ERK pathway signaling by using psMEK1 and an ERK kinase translocation reporter with a mRuby2 fluorescent reporter (ERK KTR-mRuby2).
The ERK KTR is a target for MEK1 phosphorylation, much like endogenous ERK, and its localization serves as an indicator of MEK1 activity. When MEK1 is active, it phosphorylates the ERK KTR and red fluorescence localizes to the cytoplasm. When MEK1 and ERK are inactive, the ERK KTR isn’t phosphorylated and red fluorescence can be found in the nucleus. For testing kinase inhibitors, psMEK1 and ERK KTR-mRuby2 expressing cells were incubated with an inhibitor of choice, exposed to 500 nm cyan light to activate psMEK1, and the localization of ERK KTR-mRuby2 determined. If the inhibitor targeted MEK1 or its downstream target ERK, the KTR remained in the nucleus. But if the inhibitor targeted upstream members of the pathway (Raf1, EGRF), the KTR localized to the cytoplasm.
 This all-optical psMEK1 + ERK KTR-mRuby2 system has several advantages over existing methods. First, it avoids activating other signaling pathways, which frequently occurs when serum or growth factors are used as a stimulus. This system also allows precise modulating of a particular arm of the signaling pathway (i.e. everything downstream of MEK1), bypassing the upstream portion of the pathway and focusing on the downstream steps. It’s also possible to determine immediate effects of signaling vs the delayed effects because you can precisely control the timing of kinase activity.
This all-optical psMEK1 + ERK KTR-mRuby2 system has several advantages over existing methods. First, it avoids activating other signaling pathways, which frequently occurs when serum or growth factors are used as a stimulus. This system also allows precise modulating of a particular arm of the signaling pathway (i.e. everything downstream of MEK1), bypassing the upstream portion of the pathway and focusing on the downstream steps. It’s also possible to determine immediate effects of signaling vs the delayed effects because you can precisely control the timing of kinase activity.
2. Studying negative feedback loops of signaling pathways
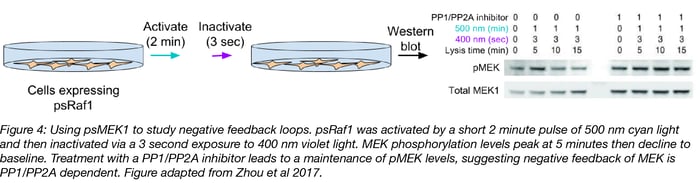
Negative feedback loops are a common feature of signal transduction pathways, but are often hard to decipher because it’s difficult to create short pulses of kinase activity. Zhou et al studied the negative feedback on endogenous MEK1/2 phosphorylation by creating a short pulse of psRaf1 activity. They did this by exposing psRaf1 to a short 2 minute pulse of 500 nm light (turn on psRaf1) followed by 3 seconds of 400 nm light (turn off psRaf1). After this, the phosphorylation status of MEK1/2 was tracked at 5 minute intervals via Western blot. Phosphorylation of MEK1/2 peaks at just 5 minutes post-Raf1 inactivation, and then decreases. Treatment of the cells with a PP1/PP2A inhibitor leads to maintenance of phosphorylated MEK1/2 levels, suggesting PP1/PP2A is responsible for this MEK1/2 negative feedback.
3. Studying signaling pathways in vivo
 Expressing photoswitchable kinases in vivo not only allows for precise control of a signaling pathway in the context of a whole organism, but also allows for precise activation of a kinases in a body part of choice. Zhou et al again used the Raf-MEK-ERK signaling pathway to demonstrate the utility of photoswitchable kinases in vivo with C. elegans.
Expressing photoswitchable kinases in vivo not only allows for precise control of a signaling pathway in the context of a whole organism, but also allows for precise activation of a kinases in a body part of choice. Zhou et al again used the Raf-MEK-ERK signaling pathway to demonstrate the utility of photoswitchable kinases in vivo with C. elegans.
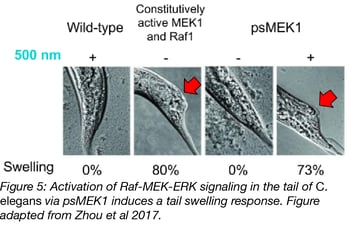
Researchers commonly use rectal bacterial infection to induce tail swelling in C. elegans, but activation of Raf-MEK-ERK signaling in the hypodermis and gut cells of C. elegans causes the same phenotype. Activation of the Raf-MEK-ERK signaling pathway with psRaf1 or psMEK1 in these same cells by 24-48 hours exposure to blue light also induced tail swelling in ~75% of worms (see below figure). Zhou et al also used a photoswitchable CDK5 in C. elegans. See figures 4B and C in Zhou et al for details on these experiments. Photoswitchable kinases could possibly be used in other model systems, but they need to be expressed in a tissue or organism that blue and violet light can penetrate.
How can I try using photoswitchable kinases in my experiments?
You can find the plasmids from Zhou et al here. In addition, it could be possible to use pdDronpa to control other kinases and proteins. Zhou et al suggest that more photoswitchable proteins could potentially be created by inserting pdDronpa domains into loops of a protein of interest. If you plan to try this, it’s important to check that the function of the protein and pdDronpa are preserved.
References
1. Zhou, Xin X., Linlin Z. Fan, Pengpeng Li, Kang Shen, and Michael Z. Lin. "Optical control of cell signaling by single-chain photoswitchable kinases." Science 355, no. 6327 (2017): 836-42. PubMed PMID: 28232577. Figures 2-4 were modified from this reference.
2. Regot, Sergi, Jacob J. Hughey, Bryce T. Bajar, Silvia Carrasco, and Markus W. Covert. "High-Sensitivity Measurements of Multiple Kinase Activities in Live Single Cells." Cell 157, no. 7 (2014): 1724-734. PubMed PMID: 24949979. PubMed Central PMCID: PMC4097317.
Additional Resources on the Addgene Blog
- Learn More about Photoactivatable Fluorescent Proteins
- Control Protein Expression with Cre-lox
- Control Protein Activity with GFP
Resources at Addgene.org
- Find KTR reporter plasmids
- Find photoactivatable fluorescent proteins in empty backbones in the Michael Davidson Collection
- Find Photoswitchable Fluorescent Proteins
Topics: Fluorescent Proteins, Other Fluorescent Protein Tools






Leave a Comment