Once upon a time, not so long ago, spCas9 was the only Cas enzyme widely available and applied by researchers for gene targeting. Fast forward a decade, and the CRISPR field has exploded with dozens of Cas enzymes and variants available. Without a comprehensive resource, it can be overwhelming to choose a Cas for your experiment. Even worse, you may end up missing an enzyme that is a better experimental fit due to this. But now, it’s time to cast aside those concerns and embrace a new resource to solve the problem: CasPEDIA, a new resource from Jennifer Doudna’s lab at the Innovative Genomics Institute.
What is CasPEDIA?
CasPEDIA is an online, searchable resource that provides summary information on Class 2 Cas enzymes. The information is organized in wiki format and includes biochemical properties, previously established uses, and intended applications. Within CasPEDIA, there is a classification system, CasID, to help facilitate direct comparison of different enzymes. CasPEDIA is meant to be a constantly updating database which serves as a resource for the scientific community.
What are the features?
CasPEDIA has collected relevant information on all the Cas enzymes you know and love, and even some you’ve likely never heard of. Each entry displays the following information for a Cas enzyme:
- Nuclease properties (CasID): Cis/trans activity, PAM requirement, gRNA, and multiplex ability
- Experimental guidelines and applications
- Protein structure and properties
- Links to relevant resources
The homepage displays a graphic describing CasIDs and relevant properties of Cas enzymes. That’s a lot of helpful information! To get you started, we’ll give a quick overview of each CasPEDIA feature.
Activity features: CasID
CasPEDIA will display both the cis and trans activity of each entry. For example, SpyCas9a (one of the most frequently used Cas enzymes) cleaves both strands of its dsDNA target in cis and has no trans activity. AsCas12a, on the other hand, generates staggered DNA nicks at its target in cis and cleaves ssDNA in trans. In sum, CasPEDIA documents the target substrate (DNA/RNA and strandedness) along with its action on the substrate (nick, bind, cleave, mixed). This information represents the ‘big picture’ features of the enzymes documented that should narrow the field for a candidate Cas for your experiment.
Target requirements
As we all know, a Cas guided by a gRNA is useless without an adjacent PAM or protospacer-flanking sequence (PFS) in some cases. The resource displays these requirements in regard to their sequence (NGG, etc.) as well as their orientation relative to the gRNA (5’ or 3’). In some cases, there is no requirement, which is also documented.
Experimental design
When you are in the design phase of your experimental planning, you need more nitty-gritty details — how can I multiplex this enzyme? What other RNA components are required? CasPEDIA provides information on crRNA and tracrRNA requirements, along with any other required non-CRISPR-associated host factors required for CRISPR processing. For each enzyme, it will also show you how multiplexed arrays would be designed, if that’s your end goal.
CasPEDIA also features useful links to Addgene resources throughout the site. In the Experimental Considerations section on each enzyme page, you'll find a link to search Addgene's collection for plasmids containing that enzyme, and embedded in the References lists on every page you can find direct links to plasmids from the article if they're available through Addgene. So whether you're looking for the exact Cas9 plasmid used by Jinek et al. 2012, or browsing for the latest AsCas12a plasmids, Addgene and CasPEDIA have got you covered!
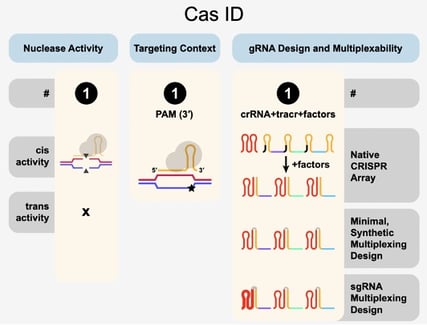
|
| Fig. 1 – CasPEDIA Cas ID display information for a given Cas (SpyCas9a) from CasPEDIA. |
Helpful resources and information
For some researchers, the previous information may be all you need to start your targeting experiment. However, many situations could arise where you still need more information. If you are performing a viral delivery of Cas, the size of the protein would be helpful to know for viral packaging limitations. Maybe you want to read literature on the less common applications of an enzyme for your non-traditional experiment. Perhaps you are a structure junky who just wants to learn about Cas domains. CasPEDIA either directly has this information or links out to this content and much, much more.
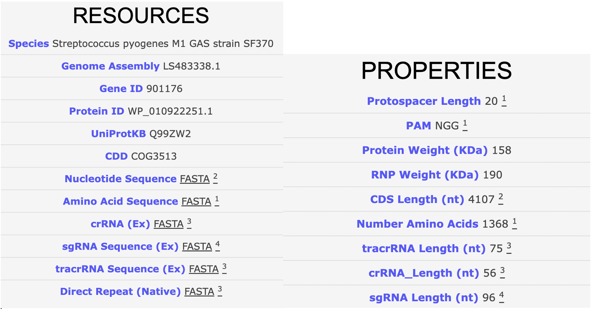
|
| Fig. 2 – List of resources and properties for a Cas entry (SpyCas9a) from CasPEDIA. |
How can I use it?
CasPEDIA operates like many search engines, featuring both a general search bar tool and an advanced search option. For the former, you simply type in identifier keywords (Cas name, RefSeq ID, etc.) and you will see a table of protein entry results. For the advanced search options, this is where you can really identify the best Cas for the job! There are a series of drop-down menus in which you can manually enter details to refine your search (trans activity, multiplex ability, gRNA design, etc.). With this feature, you can compare different Cas enzymes to make sure you are getting the desired features and cutting (note: spCas9 isn’t always the winner!).
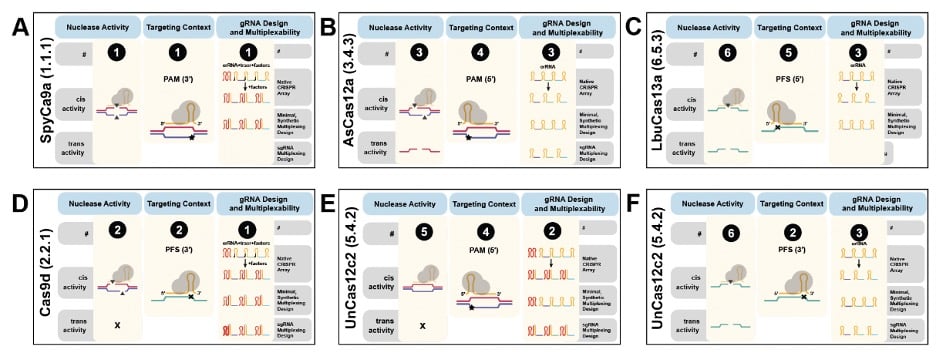 |
| Fig. 3 – Comparison of different Cas enzymes using CasPEDIA. Adapted from Adler et al. |
You can also search by protein sequence with BLAST, using the homepage search bar. Once you have your hits, you can easily sort them by other parameters, such as E-value for protein sequence searches. Additionally, the Phylogeny tool allows users to browse entries by the current phylogenetic nomenclature and discover orthologs with similar properties (Makarova et al).
CasPEDIA is an evolving database, and all researchers are invited to contribute to it! The CasPEDIA Consortium will, of course, make efforts to keep it updated with new entries and update entries as research progresses, but you are invited to reach out to suggest new entries or even new features to the tool.
Resources and References
References
Adler, B., Trinidad, M., Bellieny-Rabelo, D., et al. (2023). CasPEDIA Database: A Functional Classification System for Class 2 CRISPR-Cas Enzymes [Preprint]. Biochemistry, Biophysics, and Structural Biology. https://doi.org/10.32942/X2C31F
Makarova, K. S., Wolf, Y. I., Iranzo, J., et al. (2019). Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nature Reviews Microbiology, 18(2), 67–83. https://doi.org/10.1038/s41579-019-0299-x
Resources on Addgene.org
- CRISPR software and resources
- Addgene CRISPR Guide
- Addgene’s CRISPR 101 eBook
More resources on the Addgene blog
- CRISPR 101: Which Cas9 do I choose for my CRISPR experiment?
- CRISPR 101: Cas9 The Other Cas(s)
- The PAM Requirement and Expanding CRISPR Beyond SpCas9
More resources from the Innovative Genomics Institute
Topics: CRISPR





Leave a Comment