Did you catch our April AAV webinar with Tim Miles, PhD, Director of the CLOVER Center at CalTech? If so, you may have submitted a question that didn’t get answered live - but he kindly took some time to address all your unanswered questions via text! (well, maybe not all of them. We still haven’t gotten to the bottom of why hot dogs are sold in packages of 8 yet hot dog buns are sold in packages of 10…)
If you missed the webinar, you can watch the presentation and Q&A session on our YouTube channel. And with no more ado, the questions!
 AAV Prep and Technique
AAV Prep and Technique
Q1: Is there any way to deliver a long gene (>4.5 kB) by AAV?
A: One potential strategy is to use split-inteins to mediate trans-protein splicing. Note, however, that this will require co-delivery of two AAVs at a high MOI, often resulting in a drop in transduction coverage compared to just one vector.
Q2: Do you think it is possible to make an efficient AAV prep for a 4.2 kB insert with the CMV promoter using the AAV-BR1 capsid?
A: It is our understanding that AAV-BR1 has the same genome size restrictions as other AAV. Therefore, one should aim to keep their cargo below 4.4kB (with the ITRs bringing total genome length to ~4.7kB. If your insert alone is 4.2 kB and CMV promoter+enhancer is 0.5 kB, then you would be oversized even before considering the need for some manner of polyA. In this scenario, you may be best served by a dual vector, split intein approach.
Q3: What’s the best method for titering AAV? Is there a difference in titer results between ddPCR and qPCR?
A: At CLOVER, we have traditionally used qPCR (protocol link). We have found this method to be reliable and reproducible. However, we do not use amplicons in ITR as we have found this to strongly bias titer. We are also adopting ddPCR, which does not a standard curve and somewhat more reliable quantitation. Differences in sample treatment across methods can affect reported titer by either method so to ensure equal animal dosing, we advise that users keep titering method consistent across samples within a project.
Targeting Your AAV
Q4: Are there any AAVs that have successful transduction in microglia? I have been using the AAV.PHP.eb for neuron, oligodendrocyte, and astrocyte specificity and would like to get it working in microglia as well.
A: We are not aware of any available AAV for efficiently targeting microglia in vivo.
Q5: What is the efficiency of co-delivering different AAV particles into the same cells in vivo?
A: This will be highly dependent on vector dose. The color diversity in Vector Assisted Spectral Tracing (VAST) experiments using PHP.eB in the brain give some insight into codelivery efficiency at different doses. See Figure 4 in this paper for more information.
Q6 & Q7: We have been attempting to transduce mouse liver via tail vein injections but would like to avoid "leaking" into the brain, would you recommend using the PHP.S capsid over the PHP.eB (the capsid we have been using)?
Would you recommend any liver specific capsid, particularly one that can target the epithelial cells in the blood-brain barrier?
A: For liver, we recommend using one of the natural serotypes such as AAV9 or AAV8. Very recently an engineered AAV5 capsid was reported for specific and potent liver transduction. While we have not tried it, it might be worth investigating for both Q6 & Q7.
For epithelial cells of the blood brain barrier we would currently recommend PHP.V1 or AAV-BR1. We recommend you keep an eye out for emerging new capsids in this area.
Q8: On dense labeling in the brain with PHP.eB: A recent CLOVER collaboration injected 1e+13 vg per mouse to get phenotypes (that’s almost 100ul/animal for the PHP.eBs that we have in house!), which seems to me a very high dosage. Any thoughts?
A: The question refers to this paper. AAV can be safely concentrated to ~1x1014 vg/ml.. As evident in our repository, this application is an outlier for PHP.eB (1x1011 - 1x1012 vg/mouse is more standard) and reflects a desire for very high MOI on top of very high coverage. Please see this paper for more information about PHP.eB brain labeling at various doses.
As always, you will need to discuss injection volumes with your IACUC committee.
Animal Models
Q9: Is there any difference in transfection efficiency when delivering these viruses via retro-orbital vs. tail vein systemic injections?
A: We have not observed any appreciable difference in AAV transduction efficiency between retro-orbital versus tail vein injections.
Q10: Have you or others been able to successfully systemically transduce and then retransduce a model organism such as mice, e.g. first at 2 months, then at 4 months? Or do antibodies become a prohibitive problem?
A: Re-administration is a challenge for AAV due to the development of neutralizing antibodies in animal models. All of our engineered AAV are presently built on an AAV9 backbone and so will very likely cross-react with each other. Multiple labs have tried to transfer PHP.eB's enhanced CNS tropism to other serotypes, but reproducible success remains elusive.
There has also been a lot of effort in immunomodulation or immune suppression to allow AAV re-administration but I do not think the field has settled on ‘best practices’ for model organisms.
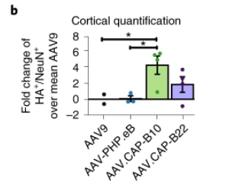 Q11: How consistent is expression is when injecting CAP-B10 or CAP-B22 in marmosets, from an individual to another individual perspective?
Q11: How consistent is expression is when injecting CAP-B10 or CAP-B22 in marmosets, from an individual to another individual perspective?
A: CAP-B10 and CAP-B22 were tested individually in 4 marmosets each. Brain potency for each individual is reported in Figure 4B (see left for image and link for paper.)
Q12: Is there any AAV serotype that can be used in Drosophila?
A: Not to our knowledge.
Disease Applications
Q13: Does AAV have potential applications in diseases which may be linked to epigenetic changes, such as multiple sclerosis?
A: Basic science and preclinical researchers have used the flexibility of AAV genome design in remarkably creative ways. The continued development of more and smaller CRISPR tools is an exciting area for future AAV application to epigenetic editing. This area is discussed in our recent review.
And that's a wrap on our AAV webinar! Special thanks to our panel of experts, Tim Miles, David Goertson, Xinhong Chen, and Miggy Chuapoco, and to all our participants for their time and excellent questions.
Additional Resources on the Addgene Blog
- Using virus in your research - a primer for beginners
- Adenoviral delivery of CRISPR/Cas9
- Tips for a first time AAV user
Resources on Addgene.org
Topics: Viral Vectors, AAV, Adenoviral Vectors, Q&A







Leave a Comment