 Every quarter we highlight a subset of the new plasmids in the repository through our hot plasmids articles. These brief articles provide a synopsis of a plasmid or group of plasmids' functions and applications. We hope that these articles make it easier for you to find and use the plasmids you need. You can find all the hot plasmids from 2017 below. With over 50,000 plasmids, we can't write posts for every great plasmid that comes into the repository, but be sure to let us know if you'd like to write about your plasmids in a future blog post. No time to read?
Every quarter we highlight a subset of the new plasmids in the repository through our hot plasmids articles. These brief articles provide a synopsis of a plasmid or group of plasmids' functions and applications. We hope that these articles make it easier for you to find and use the plasmids you need. You can find all the hot plasmids from 2017 below. With over 50,000 plasmids, we can't write posts for every great plasmid that comes into the repository, but be sure to let us know if you'd like to write about your plasmids in a future blog post. No time to read?
Listen to our hot plasmids segment on the Addgene Podcast.
Subscribe to the Addgene blog to get all the latest hot plasmid updates!
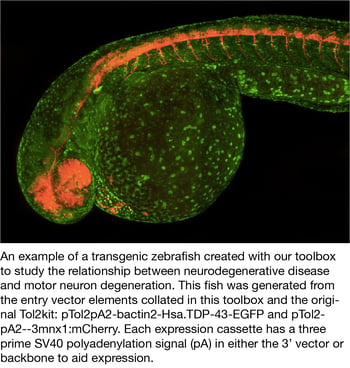
Tol2 Gateway-compatiable toolbox for studying the nervous system
The Cole lab recently deposited the Tol2 Gateway-Compatible Toolobox, which will allow scientists to quickly and easily generate zebrafish transgenic lines to study neurobiology and neuorodgenerative diseases.
The Tol2 Gateway-Compatible Toolbox is based on the original Tol2kit generated by the Chi-Bin Chien lab (Kwan et al., 2007) and includes four promoters, six fluorophores with nonoverlapping emission spectra (N- and C-terminal tags for mTagBFP, TagRFPt, EGFP, mVenus, mCerulean3, mKOFP2) and empty vectors that have standard cloning sites or gateway compatible cloning sites for easy cloning of your genes of interest. In order to improve the study of specific neuronal cell types involved in neurodegenerative diseases, the Cole Lab selected promoters for cell types directly linked to disease, including motor neurons (-3mnx1/Hb9), pan neuronal (elavl3/HuC), microglial/macrophage (mpeg1.1), and astrocytic (gfap).This toolbox adds new neuronal tools to the expanding gateway compatible vectors currently shared amongst the zebrafish community.
- Don E, et al. Zebrafish. 2017. PubMed PMID: 27631880
- Kwan K, et al. Dev Dyn. 2007. PubMed PMID: 17937395
Listen to Our Tol2 Gateway-Compatible Toolbox Podcast Segment
The impact of KLF4 N-terminal variants on iPSC generation
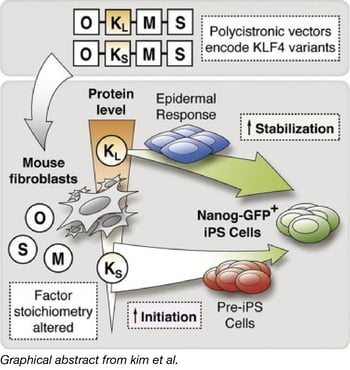 Deriving iPSCs (induced pluripotent stem cells) through the expression of transcription factors OCT3/4, SOX2, KLF4, and c-MYC is the established model for reprogramming somatic cells. A common method for generating iPSCs to express the transcription factors from a polycistronic cassette. However, it was unknown whether variations in gene expression from these polycistronic cassettes could lead to comparable experimental results.
Deriving iPSCs (induced pluripotent stem cells) through the expression of transcription factors OCT3/4, SOX2, KLF4, and c-MYC is the established model for reprogramming somatic cells. A common method for generating iPSCs to express the transcription factors from a polycistronic cassette. However, it was unknown whether variations in gene expression from these polycistronic cassettes could lead to comparable experimental results.
Using PiggyBac transposons, the Woltjen Lab compared different variants of the polycistronic cassettes and discovered reprogramming discrepancies, which they traced to a 9 amino acid N-terminal variation in the Klf4 isoform (KLF4s or KLF4L). Polycistronic cassettes that contained KLF4s had overall diminished KLF4 expression levels; this altered stoichiometry impacted reprogramming and global gene expression patterns. Polycistronic cassettes that contained the KLF4L isoform had more robust initiation of reprogramming (evaluated by an alkaline phosphatase-positive assay) as well as stabilization (Nanog-GFP reporter activation and silencing of factor-linked mCherry). They additionally discovered that monocistronic expression of either variation of Klf4 did not lead to the observed reprogramming discrepancies.
The polycistronic piggyBac transposon vectors tested in this work are available at Addgene.
- Kim, et al. Stem Cell Reports. 2015. PubMed PMID: 25772473
Targeting and manipulating interneurons using a viral approach
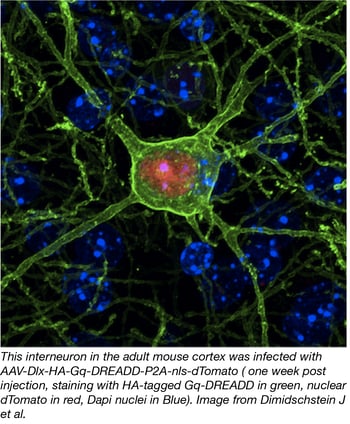
Genetically modified organisms like transgenic mice are important tools for understanding the function of cell types in the nervous system. While certain cell types, such as principal excitatory cells and glia, have been successfully targeted via viral particles, an effective approach to target specific neuronal subtypes was not available until now.
In order to target specific neuronal subtypes such as inhibitory GABA-ergic interneurons, Gordon Fishell’s lab has developed a new AAV approach using regulatory elements that restrict expression to this cell type. To this end, they constructed AAV plasmids bearing one of the distal-less homeobox 5 and 6 (Dlx5/6) enhancer elements and combined it with a variety of reporters (i.e. eGFP) and effectors (i.e. DREADDs). The team demonstrated that viral expression is robust and specific to interneurons in mice as well as other organisms, opening the possibility of using these tools in virtually any vertebrate.
This novel AAV approach allows you to target and functionally manipulate GABA-ergic interneurons across species and will undoubtedly constitute a valuable tool for the research community.
You can find the AAV vectors for infecting GABA-ergic interneurons here.
- Dimidschstein J, et al. Nature Neuroscience. 2016. PubMed PMID: 27798629
Listen to Our Podcast Segment on This New AAV Tool
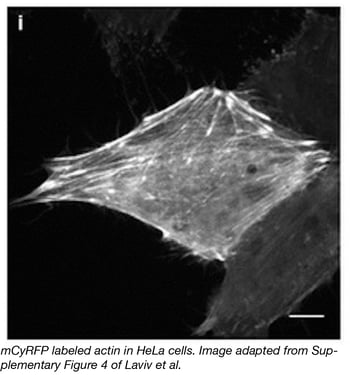 Researchers in the labs of Michael Lin and Ryohei Yasuda have teamed up to develop a novel monomeric cyan-excitable red fluorescent protein (mCyRFP1). Derived from cyan-excitable organge fluorescent protein (CyOFP1), some of the exciting properties of mCyRFP1 include a redshifted-emission spectrum compared to that of CyOFP1, a large stokes shift, long fluorescence lifetime, and a stable monomeric form as determined by the organized smooth endoplasmic reticulum (OSER) assay. A major advantage of mCyRFP1 is its ability to be coexcited along with EGFP while retaining an emission spectrum that is easily separable from that of EGFP. This makes mCyRFP1 a powerful tool for use as a donor in FRET (fluorescence resonance energy transfer) and FLIM (Fluorescence Lifetime Imaging Microscopy) applications. To demonstrate the power of mCyFP1, the Lin and Yasuda groups use both mCyFP1 and EGFP as FRET donors in experiments simultaneously measuring the activities of CaMKII and RhoA during induction of structural plasticity in single dendritic spines. The set of CyRFP constructs available at Addgene includes bacterial and mammalian mCyRFP1 or CyRFP1 expression plasmids, FRET constructs, and CaMKII and RhoA sensors.
Researchers in the labs of Michael Lin and Ryohei Yasuda have teamed up to develop a novel monomeric cyan-excitable red fluorescent protein (mCyRFP1). Derived from cyan-excitable organge fluorescent protein (CyOFP1), some of the exciting properties of mCyRFP1 include a redshifted-emission spectrum compared to that of CyOFP1, a large stokes shift, long fluorescence lifetime, and a stable monomeric form as determined by the organized smooth endoplasmic reticulum (OSER) assay. A major advantage of mCyRFP1 is its ability to be coexcited along with EGFP while retaining an emission spectrum that is easily separable from that of EGFP. This makes mCyRFP1 a powerful tool for use as a donor in FRET (fluorescence resonance energy transfer) and FLIM (Fluorescence Lifetime Imaging Microscopy) applications. To demonstrate the power of mCyFP1, the Lin and Yasuda groups use both mCyFP1 and EGFP as FRET donors in experiments simultaneously measuring the activities of CaMKII and RhoA during induction of structural plasticity in single dendritic spines. The set of CyRFP constructs available at Addgene includes bacterial and mammalian mCyRFP1 or CyRFP1 expression plasmids, FRET constructs, and CaMKII and RhoA sensors.- Laviv, et al. Nat Methods. 2016. PubMed PMID: 27798609
Listen to our mCyRFP1 Podcast Segment
New bright monomeric red fluorescent protein - mScarlet!
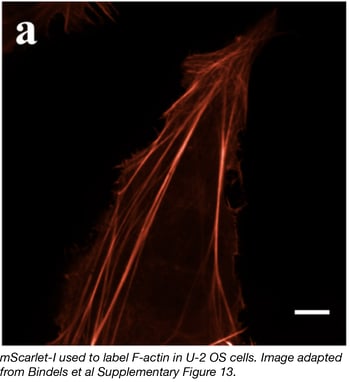 The Dorus Gadella lab has recently developed a synthetic red fluorescent protein variant they call mScarlet. Bindels et al used a combination of deliberate and random mutagenesis to isolate the best RFP. They targeted known residues on the outer surface to break dimerization interfaces (affecting many of the RFPs developed from natural ancestors), and screened through libraries of new RFP variants for increased fluorescence, brightness, and proper maturation. Three new RFPs were isolated and characterized: mScarlet (fluorescent lifetime of 3.9 ns, quantum yield of 0.70), mScarlet-I with T74I (accelerated maturation in cells compared to mScarlet, fluorescent lifetime of 3.1 ns, quantum yield of 0.54) and mScarlet-H with M164H (2-fold improvement in photostability compared to mScarlet, fluorescent lifetime of 1.3 ns, quantum yield of 0.20). All three mScarlet variants have shown great performance in cellular functional imaging and in protein fusions. The Gadella lab has deposited all mScarlet constructs with Addgene.
The Dorus Gadella lab has recently developed a synthetic red fluorescent protein variant they call mScarlet. Bindels et al used a combination of deliberate and random mutagenesis to isolate the best RFP. They targeted known residues on the outer surface to break dimerization interfaces (affecting many of the RFPs developed from natural ancestors), and screened through libraries of new RFP variants for increased fluorescence, brightness, and proper maturation. Three new RFPs were isolated and characterized: mScarlet (fluorescent lifetime of 3.9 ns, quantum yield of 0.70), mScarlet-I with T74I (accelerated maturation in cells compared to mScarlet, fluorescent lifetime of 3.1 ns, quantum yield of 0.54) and mScarlet-H with M164H (2-fold improvement in photostability compared to mScarlet, fluorescent lifetime of 1.3 ns, quantum yield of 0.20). All three mScarlet variants have shown great performance in cellular functional imaging and in protein fusions. The Gadella lab has deposited all mScarlet constructs with Addgene.
- Bindels DS, et al. Nat. Methods. 2017. PubMed PMID: 27869816
Listen to our mScarlet Podcast Segment
Photocleavable protein PhoCl expands optogenetics toolbox
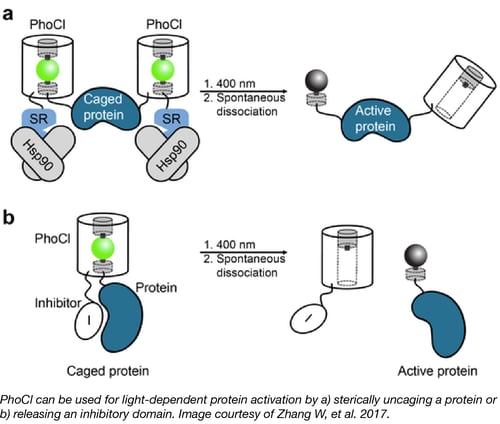
Increasingly popular Optogenetics tools use light to alter cell physiology and molecular processes via genetically encoded proteins. PhoCl is a new member of this class that works via a different mechanism - the PhoCl protein is irreversibly cleaved when exposed to light.
Robert Campbell’s lab illustrates the utility of PhoCl in a variety of settings. For instance, inserting PhoCl in between a protein and a tag, such as an NLS or NES, enables control over localization. Inserting PhoCl between an enzyme and an inhibitor domain similarly allows for control of enzymatic activity. In addition, proteins can be “caged” by fusion to specific domains thereby rendering them inactive in the cytoplasm. The authors use PhoCl in combination with this caging technique to beautifully demonstrate light-dependent Gal4 transcriptional activation and Cre recombination.
Find PhoCl plasmids at Addgene.
- Zhang W, et al. Nature Methods. 2017. PubMed PMID: 28288123
Listen to Our Podcast Segment on PhoCl
LSSmCherry1 & RDSmCherry1: Engineering and directed evolution of new RFP variants
The toolbox of fluorescent proteins (FP) for cellular imaging is constantly expanding. Case in point - check out the growing list of popular plasmids in Addgene’s FP collection. Just last month, Robert Campbell’s lab added two new mCherry variants to the repository. These variants can be used as tools to learn about the influence of structure on an FP’s properties. pBAD-LSSmCherry1 is a long Stokes shift variant, which could be compatible with two-photon microscopy using Ti-Sapphire lasers. pBAD-RDSmCherry1 is a red-shifted variant which, with further development, could be better suited for deep-tissue in vivo imaging. These variants were developed as part of a larger strategy to engineer improved red fluorescent proteins (RFPs) and, while the authors note that there are other RFP variants available that may be preferred for their intended uses, they provide a well-defined framework for the production of additional variants. Read the group’s PLOS ONE paper for more on the directed evolution of mCherry and the spectral properties of various RFPs.
For tips on how to choose the right FP for your next experiment, check out our recent guest blog series: A Practical Approach to Choosing the B(right)est Fluorescent Protein.
- Shen Y, et al. PLoS ONE. 2017. PubMed PMID: 28241009
CRISPR coselection with ouabain increases editing efficiency
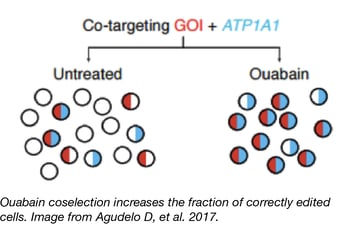 Blue flame depositor Yannick Doyon’s lab has developed a new method to increase CRISPR editing efficiency in mammalian cells. Their marker-free coselection, analogous to methods previously used in C. elegans, targets the sodium/potassium pump ATP1A1 in addition to a researcher’s locus of interest. NHEJ- or HDR-induced mutations in ATP1A1 can render cells resistant to the small molecule ouabain - and editing at the ATP1A1 locus increases the probability that the locus of interest has also been edited! Thus, ouabain selection enriches the population for CRISPR edits and improves editing efficiency. Importantly, the point mutations created by Agudelo et al. prevent ouabain binding to ATP1A1 but do not compromise the ion pump’s function or cause cellular growth delays.
Blue flame depositor Yannick Doyon’s lab has developed a new method to increase CRISPR editing efficiency in mammalian cells. Their marker-free coselection, analogous to methods previously used in C. elegans, targets the sodium/potassium pump ATP1A1 in addition to a researcher’s locus of interest. NHEJ- or HDR-induced mutations in ATP1A1 can render cells resistant to the small molecule ouabain - and editing at the ATP1A1 locus increases the probability that the locus of interest has also been edited! Thus, ouabain selection enriches the population for CRISPR edits and improves editing efficiency. Importantly, the point mutations created by Agudelo et al. prevent ouabain binding to ATP1A1 but do not compromise the ion pump’s function or cause cellular growth delays.
With eSpCas9(1.1) and coselection, Agudelo et al. achieved editing rates as high as 83% with NHEJ and 40-50% with HDR. Coselection efficiency is so high that some applications may permit working with edited “pools” of cells, eliminating the time-consuming step of single-cell isolation. Agudelo et al. also showed high editing efficiencies with AsCpf1, indicating that ouabain coselection is a robust and flexible way to increase CRISPR editing efficiency across multiple CRISPR enzymes.
Plasmids for CRISPR coselection are available at Addgene, and the Doyon lab has also provided a detailed protocol at Protocol Exchange. Plasmid maps can be found at Addgene and in Figure S12 of the publication.
- Agudelo D, et al. Nature Methods. 2017. PubMed PMID: 28417998
Listen to Our Podcast Segment on Coselection with Ouabain
New CRISPR base editors
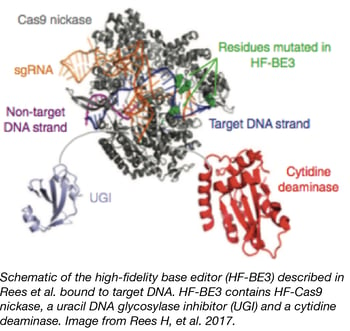 The popular CRISPR base editing technique uses dCas9/Cas9 nickase fused to a cytidine deaminase for targeted conversion of cytosine to thymine without a double stranded break. However, the method was initially limited by off-target effects, including the conversion of other cytosines near the target and in predicted Cas9 off-target sites. Two recent papers from blue flame depositor David Liu’s lab increase both the range and specificity of base editing.
The popular CRISPR base editing technique uses dCas9/Cas9 nickase fused to a cytidine deaminase for targeted conversion of cytosine to thymine without a double stranded break. However, the method was initially limited by off-target effects, including the conversion of other cytosines near the target and in predicted Cas9 off-target sites. Two recent papers from blue flame depositor David Liu’s lab increase both the range and specificity of base editing.
Kim et al. used natural and engineered Cas9 variants to develop five new base editors with distinct PAM sequences, expanding the number of available target sites for base editing. For each base editor, they observed editing activity with a minimum efficiency of ~50% and confirmed that the fusion protein retained the PAM properties of the individual Cas9. Kim et al. also mutagenized the cytidine deaminase portion of the base editor to create SpCas9 base editors with editing windows as small as 1-2 nucleotides!
To reduce off-target effects associated with base editing, Rees et al. created HF-BE3, a base editor containing high fidelity Cas9 variant HF-Cas9. They subsequently tested the original BE3 and HF-BE3 using a gRNA with high off-target cutting, and they found that HF-BE3 showed 37-fold less off-target editing than BE3, with only a slight reduction in on-target editing efficiency. To further improve specificity, they purified HF-BE3 protein for delivery in ribonucleoprotein particles (RNPs) to both zebrafish embryos and the mouse inner ear. These exciting papers further show the potential of base editing for precise genome modification.
| Plasmid ID | Plasmid Name | Cas9 Variant | Speacial Features |
| 85169 | pJL-SaBE3 | SaCas9 (NNGRRT) | |
| 85170 | pJL-SaKKH-BE3 | SaCas9 (NNGRRT) | |
| 85171 | pBK-VQR-BE3 | VQR-Cas9 (NGA) | Lower off-target activity than BE3 |
| 85172 | pBK-EQR-BE3 | EQR-Cas9 (NGAG) | Lower off-target activity than BE3 |
| 85173 | pBK-VRER-BE3 | VRER-Cas9 (NGCG) | |
| 85174 | pBK-YE1-BE3 | SpCas9 (NGG) | Editing window ~2 nt |
| 85175 | pBK-EE-BE3 | SpCas9 (NGG) | Editing window ~2 nt |
| 85176 | pBK-YE2-BE3 | SpCas9 (NGG) | Editing window ~2 nt |
| 85177 | pBK-YEE-BE3 | SpCas9 (NGG) | Lower on-target activity than BE3; editing window ~1-2 nt |
| 87438 | pET42b-HF-BE3 | HF-Cas9 (NGG) | Very low off-target activity; protein purification vector |
| 87439 | pCMV-HF-BE3 | HF-Cas9 (NGG) | Very low off-target activity; mammalian expression vector |
- Rees H, et al. Nature Communications. 2017. PubMed PMID: 28585549
- Kim Y, et al. Nature Biotechnology. 2017. PubMed PMID: 28191901
Listen to Our Podcast Segment on these New Base Editors
pPSU plasmids for generating DNA molecular weight markers
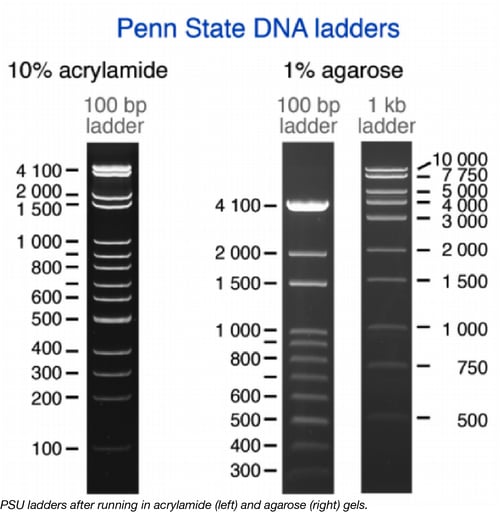
The Tan lab at Penn State - set out to “rule” Molecular Biology. Under the supervision of Dr. Tan, a team of undergraduates created DNA ladders (aka DNA molecular weight markers) that can be used to measure the size of DNA fragments. Depending on which restriction enzyme is used to cut the plasmids encoding the ladders, either 100bp or 1kb fragments are produced (see figure above).
3 Reasons The PSU Ladders Are Awesome Tools:
- They are comprised of 2 plasmids, pPSU1 and pPSU2, that can be amplified affordably in bacteria.
- They are compatible with both agarose and polyacrylamide gels.
- They are shared through Addgene without licensing restrictions.
Use these instructions to prepare the PSU ladders!
- Rosado, et al. Autophagy. 2008. PubMed PMID: 18094608
Listen to the podcast Segement on the pPSU Plasmids
B. subtilis knockout and knockdown libraries
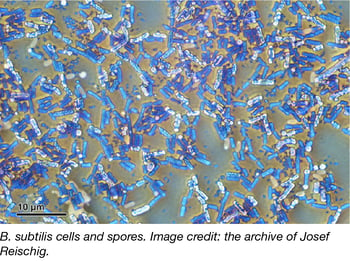
The Gross Lab at UCSF recently deposited 2 Bacillus subtilis libraries with Addgene – the B. subtilis Single Gene Deletion Library—Kanamycin and the CRISPRi Essential gene Knockdown library. Although B. subtilis has a fully sequenced genome, many applications in biotech, and is considered the most studied Gram-positive bacterium, many of its genes are poorly characterized. The Gross lab hopes these libraries will be used to advance our collective understanding of B. subtilis gene functions and interactions.
The single-gene deletion library is derived from the B. subtilis 168 strain and contains knockouts for all 3984 genes identified as non-essential. The knockouts replace entire protein-coding regions (excluding start and stop codons) with kanamycin cassettes and were carefully designed to allow for easy removal of the resistance cassette via Cre recombination. Each knockout includes unique barcodes and universal priming sites to ensure the collection is compatible with high-throughput approaches.
Since creating a knockout is not a feasible method for studying the phenotype of an essential gene, the CRISPRi Essential gene Knockdown library was constructed to allow for systematic examination of essential genes in vivo. Utilizing CRISPR interference (CRISPRi) technology, the arrayed knockdown library targets 289 genes identified or proposed to be essential in B. subtilis 168. Each individual strain includes a xylose-inducible dCas9 paired with a single, unique, and constitutively expressed sgRNA targeting the 5’ end of each essential gene.
- Koo M, et al. Cell Systems. 2016. PubMed PMID: 28189581
- Peters J, et al. Cell. 2016. PubMed PMID: 27238023
Listen to the podcast segment on the B. subtilis libraries
Transcription factor fusions elucidate iPSC reprogramming mechanism
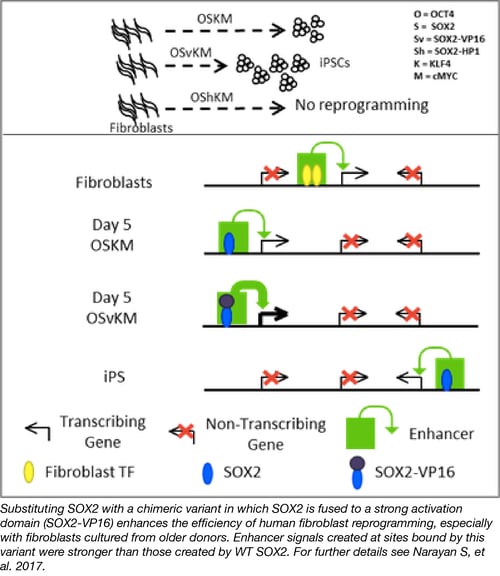
The Ptashne lab was the first to demonstrate that structurally defined domains can be swapped onto other proteins to confer functionality. In Narayan et al., they used this now classic technique to elucidate the mechanism by which SOX2 acts to reprogram human fibroblasts into induced pluripotent stem cells. By replacing wild type SOX2 with either a SOX2-VP16 (strong activator) or SOX2-HP1 (strong repressor) fusion protein, the authors show that the VP16 transcription activation domain fusion increases the efficiency of iPSC reprogramming while the HP1 fusion eliminates reprogramming. These results argue that transcription activation by SOX2 leads to its reprogramming effects.
Plasmids from the paper are available at Addgene and include a variety of cMYC, KLF4, SOX2 and OCT4 DNA binding domain and transcription activation domain fusion constructs. This is the first deposit from the Ptashne lab and we hope to see many more of their useful reagents shared via Addgene.
- Narayan S, et al. Cell Reports. 2017. PubMed PMID: 28813671
Listen to the podcast segment on modulating iPSC reprogramming
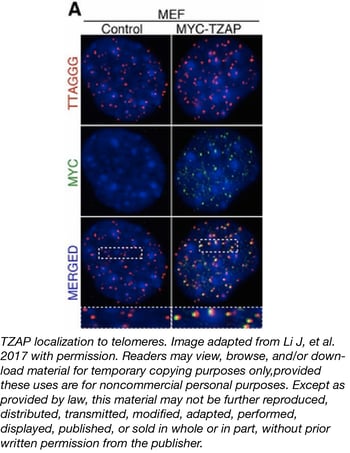
Regulation of telomere length with TZAP
Maintaining telomere length is important for cellular function and genomic stability, as telomere shortening leads to premature aging and cancer. The Denchi lab investigated an aspect of telomere length regulation called telomere trimming and have proposed a role for TZAP, a Kruppel-like zinc finger protein (previously named ZBTB48) in regulating the upper-bound of telomere length.
Telomere trimming can be assayed by looking for the production of ECT-DNA (extra-chromosomal telomeric DNA), which accumulates when T-loops (secondary telomeric structures) are deleted. Reduction in TZAP levels showed increased telomere elongation and a reduction in ECT-DNA production, while overexpression of TZAP reduced telomere length and increased ECT-DNA.
Diving further into TZAP function, Li and colleagues observed endogenous TZAP localization at telomeres and found that TZAP binds directly to TTAGGG repeats via its zinc finger domains. This binding was also independent of another TTAGGG binding complex, shelterin. Swapping the DNA binding domain of another telomeric protein, TRF2, with the zinc finger domains of TZAP showed that the TZAP zinc finger domains were sufficient to localize TRF2 to telomeres.
The plasmids used in this article have been deposited with Addgene and include MYC- and FLAG-tagged TZAP constructs (wild-type and truncation mutants), gRNAs used for TZAP gene editing, and a Hi6xs-MBP tagged TZAPznf9-11constructs used for bacterial expression and purification.
- Li J, et al. Science. 2017. PubMed PMID: 28082411
Listen to the podcast segment on regulating telomere length with TZAP
Multiplexible plant genome engineering toolkit
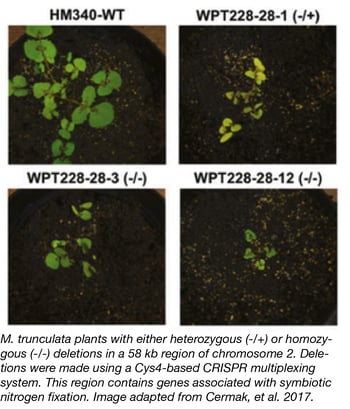 The Voytas Lab at the University of Minnesota recently deposited a comprehensive set of plasmids that can be used for either TALEN or CRISPR -based genome engineering across a variety of plant species. This toolkit includes TALEN and CRISPR nucleases, nickases, and gene activation systems as well as Cys4, tRNA, and ribozyme systems for multiplex gRNA expression. Expression of toolkit components can be controlled by a variety of promoters for versatile and robust engineering.
The Voytas Lab at the University of Minnesota recently deposited a comprehensive set of plasmids that can be used for either TALEN or CRISPR -based genome engineering across a variety of plant species. This toolkit includes TALEN and CRISPR nucleases, nickases, and gene activation systems as well as Cys4, tRNA, and ribozyme systems for multiplex gRNA expression. Expression of toolkit components can be controlled by a variety of promoters for versatile and robust engineering.
These components are provided in Golden Gate compatible backbones that can be combined with your gRNA and repair templates of choice though modular cloning steps. The Voytas lab plans to update the toolkit frequently and has developed a website to aid in plasmid design and construction. We look forward to the many plant studies that are sure to be enabled by this toolkit.
For examples of the toolkit in action, see Cermak T, et al. 2017.
- Cermak T, et al. Plant Cell. 2017. PubMed PMID: 28522548
Listen to the podcast segment on CRISPR tools for plants
CRISPR inhibitors
Bacteria utilize CRISPR Cas9 systems to defend themselves against bacteriophage infection, but some phages fight back using CRISPR inhibitors. Several such inhibitors have been characterized for type I CRISPR systems, but only recently have inhibitors for the popular type II CRISPR systems, which include S. pyogenes (Spy) Cas9, been identified.
In Rauch, et al. 2016 the Joseph Bondy-Denomy lab at UCSF identifes four proteins from Listeria monocytogenes (Lmo) prophage that can inhibit CRISPR function. These Lmo proteins were identified using a bioinformatics approach examining genomes possessing CRISPR systems with apparent self-targeting. Two of these Lmo proteins, AcrIIA2 and AcrIIA4, can work to inhibit Spy Cas9 function in both bacteria and mammalian cells.
These CRISPR inhibitors may provide a layer of regulation to genome engineering that enables new applications, such as reversing the effects of dCas9 binding to a genomic locus, or limiting the amount of time that Cas9 is active in the nucleus to reduce off-target gene editing.
The bacterial and mammalian expression vectors for these CRISPR inhibitors are listed in the table below.
| Protein | Species | Bacterial Expression Construct | SpCas9 Inhibition (E. coli) | Mammalian Expression Construct | SpCas9 Inhibition (Human Cells) |
| acrIIA1 | Listeria monocytogenes | pRAU168 | No | pJH372 | No |
| acrIIA2 | Listeria monocytogenes | pRAU171 | Partially | pJH373 | Yes |
| acrIIA3 | Listeria monocytogenes | pCSW65 | N/A (Toxic) | None | N/A |
| acrIIA4 | Listeria monocytogenes | pCSW21 | Yes | pJH376 | Yes |
| acrIIA3 | Streptococcus pyogenes | pCSW24 | N/A (Toxic) | pJH375 | No |
Topics: Addgene News







Leave a Comment