Annotation of genes in immune cells typically involves the creation of germline knockout mice, which is time-consuming, as it only changes one gene at a time. CRISPR-based systems enable gene knockout in immune cells in a high-throughput manner, but these systems have not been widely employed in vivo. The CHIME and new X-CHIME systems, developed and deposited at Addgene by Arlene Sharpe’s lab, allow for wide deployment of in vivo gene knockout in immune systems.
CHIME
We previously developed CHIME (CHimeric IMmune Editing system) to enable the constitutive, ubiquitous knockout of single genes in immune cells in vivo (LaFleur et al., 2019a and LaFleur et al., 2019b). CHIME works through transduction of Cas9-expressing (Rosa26-Cas9) hematopoietic stem cells with a lentiviral vector containing a gRNA. These transduced stem cells can be implanted in irradiated recipients to create mice with gene knockouts in their immune systems.
However, many genes have different functions in different cell types, at different times, and in combination with different genes. Having CRISPR systems for assessing genes in defined contexts would be quite useful for interrogating these scenarios.
X-CHIME
We recently developed and deposited four new systems to fill this gap. To enable context-specific knockouts, we developed X-CHIME, a suite of four systems which utilizes custom mouse strains and lentiviral expression vectors (Table 1).
The four systems
|
Name |
Function |
Plasmid |
Transduction Marker |
Mouse Strain |
|
C-CHIME |
Knocks out a pair of genes |
Violet-excited GFP (vex) |
Rosa26-Cas9 |
|
|
I-CHIME |
Inducible knockout of a single gene |
Violet-excited GFP (vex) |
Rosa26-FlpO-ERT2; H11-Cas9 |
|
|
L-CHIME |
Knocks out a single gene in defined cell types (lineage-based) |
Violet-excited GFP (vex) |
Lineage-Cre; Rosa26-Cas9 |
|
|
S-CHIME |
Knocks out pairs of genes sequentially |
Violet-excited GFP (vex) |
Rosa26-FlpO-ERT2; H11-Cas9 |
Table 1: X-CHIME systems
Proof of Concept
These systems were benchmarked on positive control genes in CD4+ and CD8+ T cells. To demonstrate the power of these systems to investigate in vivo mechanisms, we targeted two phosphatases, PTPN1 and PTPN2, which are of interest as cancer immunotherapy targets but are embryonic lethal, hindering studies post embryogenesis. Using C-CHIME and S-CHIME, we discovered that double knockout of PTPN1 and PTPN2 in immune cells in a developing or developed immune system was lethal (LaFleur et al., 2024).
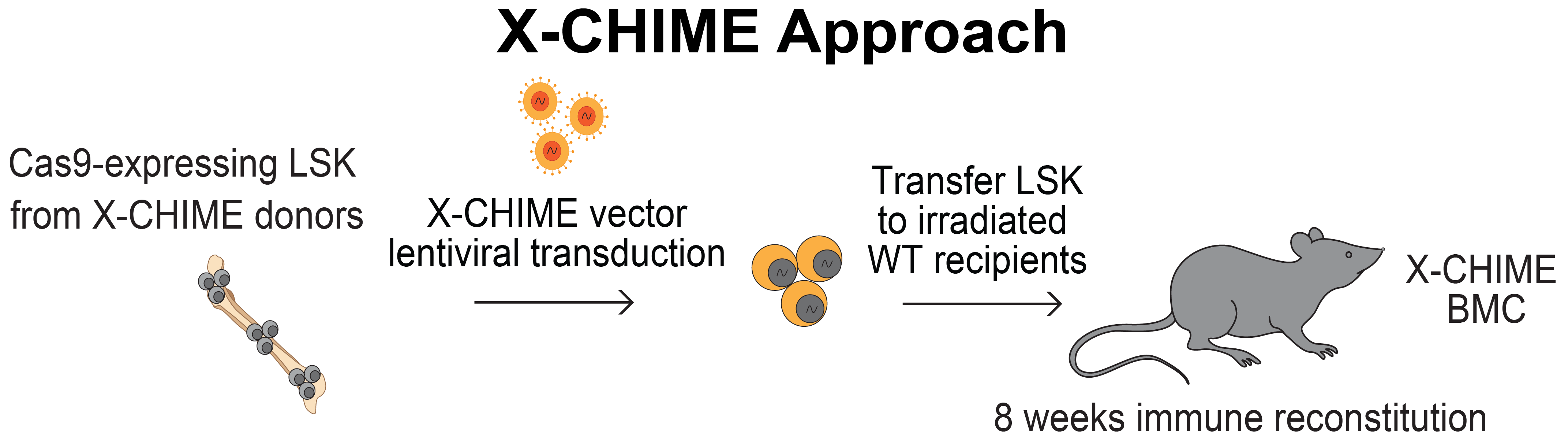 |
| Figure 1: Schematic showing the X-CHIME approach. Figure from LaFleur, et al. 2024. |
The X-CHIME deposit also includes pXPR_071, a plasmid that leads to higher titer lentivirus preparations than the first generation CHIME (pXPR_053).
-min.jpg?width=68&height=97&name=0136_Martin_Lafleur_001%20(1)-min.jpg) Marty LaFleur is a Postdoctoral Fellow in Arlene Sharpe’s laboratory at Harvard Medical School and is interested in improving CD8+ T cell responses to cancer.
Marty LaFleur is a Postdoctoral Fellow in Arlene Sharpe’s laboratory at Harvard Medical School and is interested in improving CD8+ T cell responses to cancer.
References and resources
References
LaFleur, M. W., Lemmen, A. M., Streeter, I. S. L., Nguyen, T. H., Milling, L. E., Derosia, N. M., Hoffman, Z. M., Gillis, J. E., Tjokrosurjo, Q., Markson, S. C., Huang, A. Y., Anekal, P. V., Montero Llopis, P., Haining, W. N., Doench, J. G., & Sharpe, A. H. (2024). X-CHIME enables combinatorial, inducible, lineage-specific and sequential knockout of genes in the immune system. Nature Immunology, 25(1), Article 1. https://doi.org/10.1038/s41590-023-01689-6
LaFleur, M.W., Nguyen, T.H., Coxe, M.A., Miller, B.C., Yates, K.B., Gillis, J.E., Sen, D.R., Gaudiano, E.F., Al Abosy, R., Freeman, G.J., Haining, W.N., & Sharpe, A.H. (2019). PTPN2 regulates the generation of exhausted CD8+ T cell subpopulations and restrains tumor immunity. Nature Immunology 20, 1335–1347. https://doi.org/10.1038/s41590-019-0480-4
LaFleur, M. W., Nguyen, T. H., Coxe, M. A., Yates, K. B., Trombley, J. D., Weiss, S. A., Brown, F. D., Gillis, J. E., Coxe, D. J., Doench, J. G., Haining, W. N., & Sharpe, A. H. (2019). A CRISPR-Cas9 delivery system for in vivo screening of genes in the immune system. Nature Communications, 10(1), 1668. https://doi.org/10.1038/s41467-019-09656-2
More resources on the Addgene blog
CRISPR101: Validating Your Genome Edit
How to Design Your gRNA for CRISPR Genome Editing
Resources from Addgene.org
CRISPR: Protocol for Genome Deletions in Mammalian Cell Lines
Topics: CRISPR, Other Plasmid Tools





Leave a Comment