This post was contributed by guest blogger Erik L. Snapp.
Stop using EGFP/GFP for fusion proteins! Despite multiple studies in high profile journal articles, many researchers remain unaware that EGFP/GFP is prone to forming noncovalent dimers. This property of EGFP can lead to significant artifacts.
If you're using green fluorescent protein or Enhanced Green Fluorescent Protein (GFP/EGFP) for a transcriptional reporter or as a general cytoplasmic label of cells, there's no problem. You're OK. However, if you fuse your protein of interest (POI) to GFP to study the protein's behavior in cells, in solution or something in between, you are using a tag with a serious drawback. The standard EGFP plasmid that used to be sold by Clontech and is in a freezer box in just about every lab in the world, is not inert. In all seriousness, EGFP/GFP has a real nontrivial propensity to noncovalently dimerize. That means that your POI fused to GFP or another fluorescent protein (FP) could be forming dimers in cells. Why should you care? Three simple ways a dimeric FP could ruin your day (and experiment) are listed below. Solutions to avoid these all too common issues follow.

It's a matter of concentration
Most FPs in nature are prone to dimerization (i.e. EGFP) [1, 2] or even form obligate tetramers (i.e. DsRed) [3]. This is a problem for fusion proteins. One of the major applications of FPs is to visualize the localization, dynamics, and behavior of a POI. As an investigator, you want a fusion tag to be inert, to not produce artifacts in your experiments. Considerable effort has gone into making FPs monomeric, yet many investigators remain ignorant of FP dimerization. Equally problematic, several reportedly monomeric FPs are not actually monomeric, at least in practical terms. The propensity of a particular type of molecule to form a dimer depends on its molecular affinity, termed the dissociation constant or Kd, and its concentration. The smaller the Kd, the more likely molecules of a particular type will interact. A protein with a nanomolar Kd will exist primarily as a dimer in cells while a protein with a high micromolar or low millimolar Kd is unlikely to form dimers in a cell. The Kd of EGFP is 0.11 mM [2]. Following the simple logic outlined above, you might think EGFP is unlikely to form dimers in a cell. Unfortunately, things are not quite so simple.
Concentration is the number of molecules within a volume. In many biological research situations, the relevant volume is that of the cell. For the physical behaviors associated with a molecule’s concentration to be accurately predicted, it is assumed that the molecules are homogenously distributed throughout the volume and can freely tumble in 720°. However, if the molecules are confined to a subregion of the cell and/or they cannot tumble in 720°, i.e. they are on a membrane or in an organelle, then their effective concentration will be much higher than if they are homogenously distributed and mobile throughout the cell. If two copies of a molecule are part of a single fusion protein, as in a FRET biosensor, then the local concentration of FPs around that fusion protein is very high. Real world examples of these situations in cells are described below (See Figure 1).
 |
|
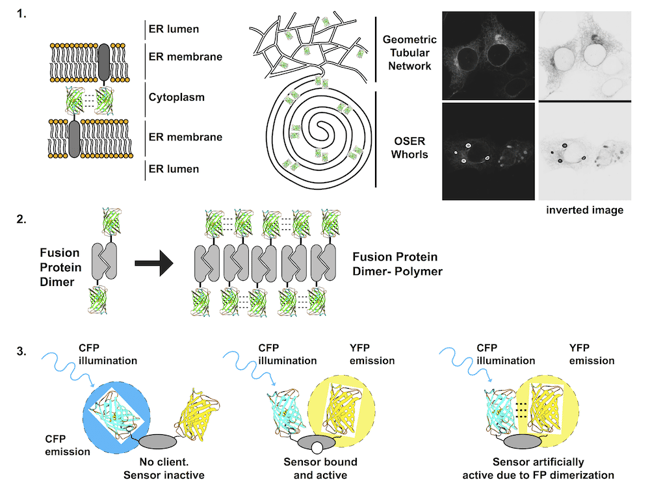
Figure 1: Each problem with dimerizing fluorescent proteins is illustrated. 1) Membrane proteins fused to FPs that dimerize can form dynamic contacts that bring apposing membranes together and can wrap the membranes into stacked structures including organized smooth endoplasmic reticulum (OSER) whorls. 2)A dimerizing protein of interest fused to a dimerizing FP has potential to form large stable polymers of dimers. 3) FRET biosensors that contain two dimerizing FPs can exist in an open inactive state, a closed active state, and an artificially active state due to closure by FP dimerization. |
1. Transmembrane fluorescent protein fusions
Transmembrane proteins, i.e. receptors and transporters, are integrated into lipid bilayers. Fusion of an FP to a transmembrane protein significantly increases the effective concentration of the FP. The FP is confined to a plane and generally only rotates in 360°, increasing the probability that two FPs will collide with each other. The consequences can be dramatic. For a membrane POI localized to the endoplasmic reticulum, the normally spiderweb-like pattern of tubules can be grossly distorted into dense stacked membranes termed Organized Smooth Endoplasmic Reticulum or OSER if a dimerizing FP is fused to the POI. These structures are large (microns), bright, non-physiological, and difficult to ignore [1, 2].
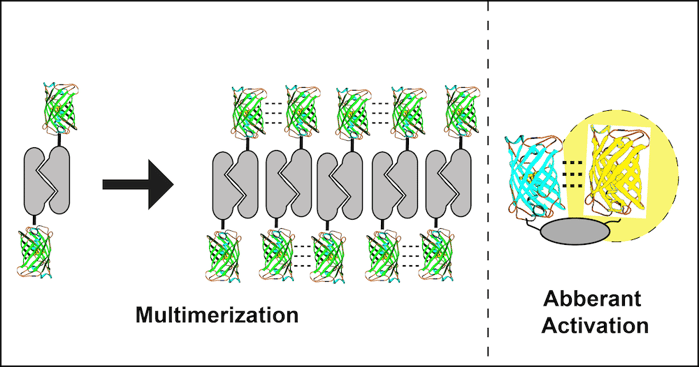
2. Fusion to an obligate dimer or oligomer
Several cell proteins normally self-associate into homodimers or even higher order oligomers. Unfortunately, FP dimerization coupled with POI dimerization can lead to polymerization of the fusion protein. A dimeric fusion protein will have two FP binding domains forming a building block sufficient to seed polymer formation as fusion protein concentration increases. The polymerized fusion proteins that form as a result will often localize incorrectly and may function improperly.
3. Incorporation into a FRET biosensor
A FRET biosensor is a reporter that undergoes a conformational change to bring two FPs on a single protein closer together or further apart. Changes in distance alter the ability of one FP to nonradiatively transfer energy to the second FP, often decreasing the fluorescent signal from the first FP and increasing the signal from the second FP. Several early FRET biosensors were made with CFP and YFP, cyan and yellow variants of EGFP equally capable of dimerizing as EGFP. As a result, CFP and YFP dimers can form in the absence of environmental changes that the biosensor is designed to detect resulting in false positives [2, 4].
Solutions to the polymer problem
There are some simple solutions available for researchers to deal with all of these problems. The simplest and best solution is to use truly monomeric FPs. EGFP and other GFP family members (superfolder, Emerald, CFP, YFP, BFP, Cerulean, Turquoise, Venus, and Citrine) can be monomerized with an A206K mutation [2]. Addgene has many of these constructs and they are designated mEGFP, mVenus, etc. A word of caution is needed because FPs not derived from GFP, i.e. the red FPs, are not necessarily monomeric, even if they were described as monomeric in a publication. The gold standard in assays used to determine if a protein is monomeric is sedimentation equilibrium analytical ultracentrifugation [2]. Some affinity assays used in the papers describing the FPs above including molecular sizing columns and native gels fail to detect EGFP dimerization. Importantly, all of these assays are performed in vitro. Assays to detect FP dimerization in in vivo cellular contexts, such as the CytERM whorl formation assay, are practical real world measures of dimerization potential that will determine if an FP will dimerize in an experimentally relevant circumstance. For a more detailed discussion see [5].
Problems 1 and 2 depend on protein concentrations. An imperfect solution is to restrict imaging analysis to cells expressing the lowest levels of the fusion protein. This may alleviate artifacts for moderately dimerizing FPs such as EGFP, but not for obligate oligomers such as DsRed or high affinity FPs such as TagRFP. Problem 3 can be solved, in most cases, by using monomeric FPs or FPs from different families, which cannot form heterodimers (i.e. EGFP and mCherry). These simple strategies can help you make better fluorescent fusion protein reporters for your studies and greatly lower your chances of finding artifactual results.
Many thanks to our guest blogger Erik L. Snapp!
 Erik Lee Snapp received his PhD in Molecular Microbiology and Immunology from Oregon Health Sciences University, did his postdoctoral fellowship with Jennifer Lippincott-Schwartz at the National Institutes of Health and he is currently an Associate Professor at Albert Einstein College of Medicine in the Department of Anatomy and Structural Biology. His interests include the quality control of secretory proteins in the Endoplasmic Reticulum and live cell imaging approaches. Erik is also an avid long distance runner, gardner, and cook.
Erik Lee Snapp received his PhD in Molecular Microbiology and Immunology from Oregon Health Sciences University, did his postdoctoral fellowship with Jennifer Lippincott-Schwartz at the National Institutes of Health and he is currently an Associate Professor at Albert Einstein College of Medicine in the Department of Anatomy and Structural Biology. His interests include the quality control of secretory proteins in the Endoplasmic Reticulum and live cell imaging approaches. Erik is also an avid long distance runner, gardner, and cook.
References
1. Snapp, E.L., et al., Formation of stacked ER cisternae by low affinity protein interactions. J Cell Biol, 2003. 163(2): p. 257-69. PubMed PMID: 14581454. PubMed Central PMCID: PMC2173526.
2. Zacharias, D.A., et al., Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science, 2002. 296(5569): p. 913-6. PubMed PMID: 11988576.
3. Matz, M.V., et al., Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol, 1999. 17(10): p. 969-73. PubMed PMID: 10504696.
4. Ohashi, T., et al., An experimental study of GFP-based FRET, with application to intrinsically unstructured proteins. Protein Sci, 2007. 16(7): p. 1429-38. PubMed PMID: 17586775. PubMed Central PMCID: PMC2206698.
5. Costantini, L.M., et al., Assessing the tendency of fluorescent proteins to oligomerize under physiologic conditions. Traffic, 2012. 13(5): p. 643-9. PubMed PMID: 22289035. PubMed Central PMCID: PMC3324619.
Additional Resources on the Addgene Blog
- Read Professor Snapp's Post on mOX FPs
- Find the Best Fluorescent Protein for Your Needs
- Get Tips for Using FRET in Your Experiments
Additional Resources on Addgene.org
- Browse our Fluorescent Protein Resources
- Find Other Plasmids that Glow (Luciferase Plasmids)
- Find Optogenetics Resources






Leave a Comment