This post was contributed by guest blogger Greg Lohman, a biochemistry researcher at New England Biolabs.
When do you need a high fidelity ligase—and when is an alternative ligase a better choice? And what is ligase fidelity anyway? Let’s talk about it.
DNA ligases are enzymes that seal breaks in DNA by joining 5 ́-phosphorylated DNA termini to 3 ́-OH DNA termini (1-4). In vitro, ligases (notably T4 DNA ligase) are critical reagents for many molecular biology protocols, including vector-insert joining for recombinant plasmid construction (restriction cloning), adaptor ligation for next generation sequencing (NGS) library construction, and circularization of dsDNA (6). Less commonly utilized in vitro, Taq DNA ligase will ligate only nicks (5-8). Taq ligase is a NAD+-dependent DNA ligase from a thermostable bacterium that can survive high temperatures (up to 95 °C) and is active over a range of elevated temperatures (37–75 °C).
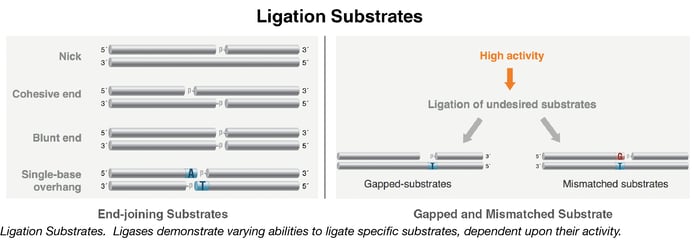
T4 DNA ligase is highly active in nick ligation and DNA end-joining, but will also efficiently ligate many undesirable structures, including substrates containing gaps or DNA base pair mismatches (8-11). For some applications, there cannot be any end-joining activity, and for others, there is a need for the exclusive ligation of fully base-paired nicks with no gaps. For example, DNA assembly methods, such as Gibson Assembly® and NEBuilder® HiFi DNA Assembly, require nick-selective ligases, such as Taq DNA Ligase, which only reacts with substrates containing no gaps, and will not join any fragments end-to-end without the exo/polymerase generation of annealed complementary regions.
Why use a high fidelity ligase?
Numerous applications have been developed to take advantage of the high fidelity of Taq and other thermostable DNA ligases. These applications detect specific nucleotide sequences with high specificity and quantitative accuracy and even include profiling single nucleotide polymorphisms (SNPs) (5,12,13). For example, in the Ligase Detection Reaction (LDR), probes complementary to the sequence of interest are annealed to target DNA (genomic DNA or a PCR amplified fragment) and treated with a high fidelity thermostable DNA ligase. If the target sequence is present, the probes will ligate. Subsequent cycling through rounds of melting and annealing allows linear amplification of the probe ligation product. With the right ligase, conditions and suitable probes, LDR can reliably detect single-base differences. The original creators of this technique visualized the ligation product in a gel, but we can now use fluorophore- quencher pairs or qPCR-based methods to greatly increase the sensitivity of detection (14-20). LDR has also been extended to multiplexed probe sets that allow the simultaneous interrogation of multiple potential SNP sites (15). The related LCR technique uses two sets of probes to achieve exponential amplification.
What does fidelity mean for a ligase?
DNA ligases generally prefer fully Watson-Crick base-paired dsDNA substrates to those containing one or more mismatches. However, some ligases can ligate mismatches to a significant degree, and very active ligases, such as T4 DNA Ligase, can ligate nicks containing one or more mismatches near the ligation junction with high efficiency (11, 21).
Ligases are thought to interrogate dsDNA for proper base pairing through minor grove contacts, and thus do not read specific base sequences, but are sensitive to distortions in helix shape (22). Large purine:purine mismatches and most smaller pyrimidine:pyrimidine mismatches are typically worse ligation substrates than pyrimidine:purine mismatches. Helix stability also plays some role, and mismatches with more hydrogen bonds are more readily ligated than those with few. For many ligases, G:T mismatches, with two hydrogen bonds and a base-pair size nearly indistinguishable from a Watson-Crick base pair, are joined with nearly the same efficiency as a correct base pair.
DNA ligases have been generally found to have a higher discrimination at the upstream side of the ligation junction (the base pair providing the 3 ́-OH terminus to the ligation) than on the downstream side (the base pair providing the 5 ́-phosphate to the ligation). The structural/mechanistic reason for this differential is not known for certain, but may have to do with the slight melting of the 5 ́-terminus during the reaction. This “peeling back” of the 5 ́-phosphorylated base can be observed in the crystal structures of several DNA ligases bound to substrate (23, 24).
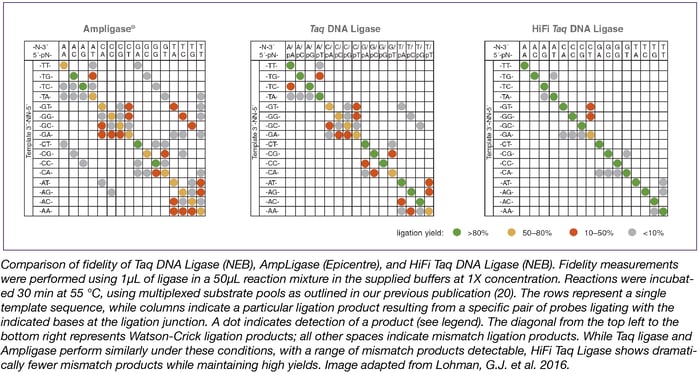
Thermostable DNA ligases, including Taq DNA ligase, are naturally able to discriminate against ligating substrates containing base pair mismatches (i.e., are "higher fidelity") better than T4 DNA Ligase (21, 25, 26). Despite this higher fidelity, Taq DNA Ligase can still detectably ligate many T:G, T:T, and A:C mismatches. Thermostable DNA ligases are active at elevated temperatures, allowing further discrimination by incubating the ligation at a temperature near the melting temperature (Tm) of the DNA strands. This selectively reduces the concentration of annealed mismatched substrates (expected to have a slightly lower Tm around the mismatch) over annealed fully base-paired substrates. Thus, high fidelity ligation can be achieved through a combination of the intrinsic selectivity of the ligase active site and careful balance of reaction conditions to reduce the incidence of annealed mismatched dsDNA.
Optimization through high-throughput profiling
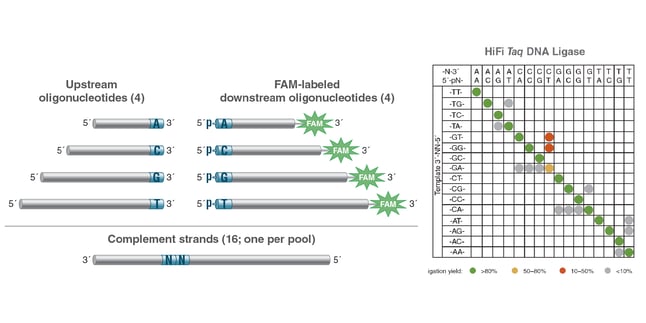
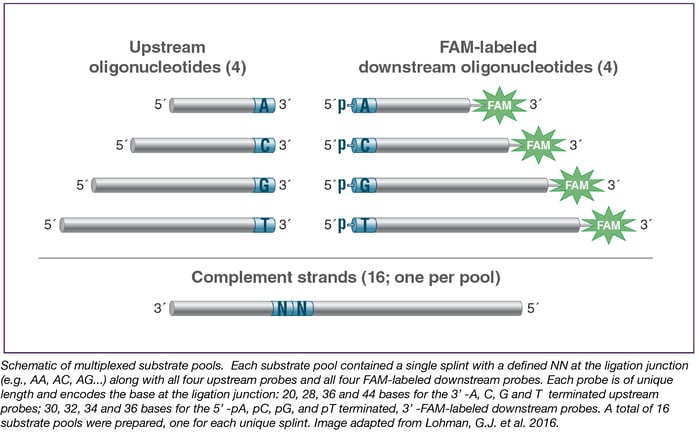
New England Biolabs® (NEB) researchers recently published a method for the high-throughput profiling of ligase fidelity that extends earlier studies through a high-sensitivity multiplexed format (21, 27). This methodology has been used to rapidly screen buffer conditions for Taq DNA Ligase and their effect on fidelity. In this method, substrate pools were prepared consisting of one target (template) strand and four upstream probes and four down-stream probes, each differing only in the base at the ligation junction. Thus, all four bases at either side of the ligation junction were represented. Sixteen separate pools were prepared, each with a different complement strand covering all 16 possible NN pairs in the template. The probes were designed such that each possible pairing resulted in a product of a different length, with products identifiable and quantifiable by capillary electrophoresis (CE) (see below). This method allowed screening of all possible base combinations (Watson-Crick and mismatched) around the ligation junction in 16 wells of a 96-well plate, allowing 6 conditions to be screened per plate. The results indicated that the optimal buffer for Taq DNA Ligase contains 100 – 200 mM KCl at pH 8.5.
This optimization method has been used internally at NEB to screen additional ligases, conditions, and formulations and has led to the development of the new HiFi Taq DNA Ligase. Using this method, both the enzyme and the reaction buffer were optimized, resulting in the highest fidelity NAD+-dependent DNA ligase commercially available (see below).
Conclusion
It is important to note that thermostable, high fidelity, nick-selective DNA ligases like Taq DNA Ligase, HiFi Taq DNA Ligase, and the ATP dependent 9°NTM DNA Ligase are not replacements for T4 DNA Ligase in applications such as routine cloning or DNA library preparation. However, when a method relies on accurate ligation of nicks lacking gaps or mismatched base pairs (as in the case for LCR, LDR and Padlock Probes), use of a high fidelity ligase, along with careful probe design and reaction condition optimization, will be critical for success. Online tools are available to help optimize ligation reactions; the Thermostable Ligase Reaction Temperature Calculator can be used to estimate an appropriate incubation temperature when using NEB thermostable ligases and reaction buffers.
Many thanks to our guest blogger, Greg Lohman!
 Greg Lohman is a biochemistry researcher at New England Biolabs, specializing in the enzymology of DNA repair enzymes.
Greg Lohman is a biochemistry researcher at New England Biolabs, specializing in the enzymology of DNA repair enzymes.
References
1. Shuman, Stewart, and Christopher D. Lima. "The polynucleotide ligase and RNA capping enzyme superfamily of covalent nucleotidyltransferases." Current opinion in structural biology 14.6 (2004): 757-764. PubMed PMID: 15582400.
2. Tomkinson, Alan E., et al. "DNA ligases: structure, reaction mechanism, and function." Chemical reviews 106.2 (2006): 687-699. PubMed PMID: 16464020.
3. Ellenberger, Tom, and Alan E. Tomkinson. "Eukaryotic DNA ligases: structural and functional insights." Annu. Rev. Biochem. 77 (2008): 313-338. PubMed PMID: 18518823. PubMed Central PMCID: PMC2933818.
4. Shuman, Stewart. "DNA ligases: progress and prospects." Journal of Biological Chemistry 284.26 (2009): 17365-17369. PubMed PMID: 19329793. PubMed Central PMCID: PMC2719376.
5. Barany, Francis. "Genetic disease detection and DNA amplification using cloned thermostable ligase." Proceedings of the National Academy of Sciences 88.1 (1991): 189-193. PubMed PMID: 1986365. PubMed Central PMCID: PMC50775.
6. Barany, Francis, and David H. Gelfand. "Cloning, overexpression and nucleotide sequence of a thermostable DNA ligase-encoding gene." Gene 109.1 (1991): 1-11. PubMed PMID: 1756968.
7. Lauer, G., et al. "Cloning, nucleotide sequence, and engineered expression of Thermus thermophilus DNA ligase, a homolog of Escherichia coli DNA ligase." Journal of bacteriology 173.16 (1991): 5047-5053. PubMed PMID: 1840584. PubMed Central PMCID: PMC208194.
8. Luo, Jianying, and Francis Barany. "Identification of essential residues in Thermus thermophilus DNA ligase." Nucleic acids research 24.15 (1996): 3079-3085. PubMed PMID: 8760897. PubMed Central PMCID: PMC146043.
9. Nilsson, Stefan V., and Göran Magnusson. "Sealing of gaps in duplex DNA by T4 DNA ligase." Nucleic acids research 10.5 (1982): 1425-1437. PubMed PMID: 7041091. PubMed Central PMCID: PMC320540.
10. Goffin, Colette, Veronique Bailly, and Walter G. Verly. "Nicks 3′ or 5′ to AP sites or to mispaired bases, and one-nucleotide gaps can be sealed by T4 DNA ligase." Nucleic acids research 15.21 (1987): 8755-8771. PubMed PMID: 3684572. PubMed Central PMCID: PMC306403.
11. Wu, Dan Y., and R. Bruce Wallace. "Specificity of the nick-closing activity of bacteriophage T4 DNA ligase." Gene 76.2 (1989): 245-254. PubMed PMID: 2753355.
12. Barany, Francis. "The ligase chain reaction in a PCR world." PCR Methods Appl 1.1 (1991): 5-16. PubMed PMID: 1842922.
13. Wiedmann, M., et al. "Ligase chain reaction (LCR)-overview and applications." PCR Methods Appl 3.4 (1994): S51-64. PubMed PMID: 8173509.
14. Zirvi, Monib, et al. "Ligase-based detection of mononucleotide repeat sequences." Nucleic acids research 27.24 (1999): e40-e47. PubMed PMID: 10572192. PubMed Central PMCID: PMC148763.
15. Pingle, Maneesh R., et al. "Multiplexed identification of blood-borne bacterial pathogens by use of a novel 16S rRNA gene PCR-ligase detection reaction-capillary electrophoresis assay." Journal of clinical microbiology 45.6 (2007): 1927-1935. PubMed PMID: 17428930. PubMed Central PMCID: PMC1933048.
16. Cheng, Changming, et al. "Electrochemiluminescent assay for detection of extremely rare mutations based on ligase reaction and bead enrichment." Analytical biochemistry 434.1 (2013): 34-38. PubMed PMID: 23142428.
17. Hamada, Mariko, et al. "Development of a ligase detection reaction/CGE method using a LIF dual‐channel detection system for direct identification of allelic composition of mutated DNA in a mixed population of excess wild‐type DNA." Electrophoresis 34.9-10 (2013): 1415-1422. PubMed PMID: 23463388.
18. Hommatsu, Manami, et al. "Development of a PCR/LDR/flow-through hybridization assay using a capillary tube, probe DNA-immobilized magnetic beads and chemiluminescence detection." Analytical Sciences 29.7 (2013): 689-695. PubMed PMID: 23842410.
19. LeClair, Norbert P., et al. "Optimization of a ligase detection reaction-fluorescent microsphere assay for characterization of resistance-mediating polymorphisms in African samples of Plasmodium falciparum." Journal of clinical microbiology 51.8 (2013): 2564-2570. PubMed PMID: 23720790. PubMed Central PMCID: PMC3719660.
20. Watanabe, Sho, et al. "Microbead-based ligase detection reaction assay using a molecular beacon probe for the detection of low-abundance point mutations." Analytical chemistry 86.1 (2013): 900-906. PubMed PMID: 24328157.
21. Lohman, Gregory JS, et al. "A high-throughput assay for the comprehensive profiling of DNA ligase fidelity." Nucleic acids research 44.2 (2015): e14-e14. PubMed PMID: 26365241. PubMed Central PMCID: PMC4737175.
22. Liu, Pingfang, Artur Burdzy, and Lawrence C. Sowers. "DNA ligases ensure fidelity by interrogating minor groove contacts." Nucleic acids research 32.15 (2004): 4503-4511. PubMed PMID: 15328364. PubMed Central PMCID: PMC516055.
23. Pascal, John M., et al. "Human DNA ligase I completely encircles and partially unwinds nicked DNA." nature 432.7016 (2004): 473. PubMed PMID: 15565146.
24. Nandakumar, Jayakrishnan, Pravin A. Nair, and Stewart Shuman. "Last stop on the road to repair: structure of E. coli DNA ligase bound to nicked DNA-adenylate." Molecular cell26.2 (2007): 257-271. PubMed PMID: 17466627.
25. Luo, Jianying, Donald E. Bergstrom, and Francis Barany. "Improving the fidelity of Thermus thermophilus DNA ligase." Nucleic acids research 24.15 (1996): 3071-3078. PubMed PMID: 8760896. PubMed Central PMCID: PMC146030.
26. Nishida, Hirokazu, et al. "Overexpression, purification and crystallization of an archaeal DNA ligase from Pyrococcus furiosus." Acta Crystallographica Section F: Structural Biology and Crystallization Communications 61.12 (2005): 1100-1102. PubMed PMID: 16511246. PubMed Central PMCID: PMC1978145.
27. Greenough, Lucia, et al. "Adapting capillary gel electrophoresis as a sensitive, high-throughput method to accelerate characterization of nucleic acid metabolic enzymes." Nucleic acids research 44.2 (2015): e15-e15. PubMed PMID: 26365239. PubMed Central PMCID: PMC4737176.
Additional Resources on the Addgene Blog
Resources on Addgene.org
Topics: Plasmid Cloning, Plasmids





Leave a Comment