This post was contributed by guest blogger Lydia Morrison from New England Biolabs.
What is DNA assembly? In the context of cloning, DNA assembly refers to a method of physically joining multiple fragments of DNA to create a synthetically designed DNA sequence. There are multiple methods of DNA assembly available, including: Gibson Assembly®, BioBrick® Assembly, Golden Gate Assembly, and NEBuilder® HiFi DNA Assembly. Gibson Assembly allows the production of scarless DNA constructs using homologous regions to guide the joining reaction. BioBrick Assembly will leave scar regions at the site of fragment joining, but this is fine for its goal of creating a choice of standardized constructs and tools for the rational and simple shuffling of DNA regions. Golden Gate Assembly also allows the creation of standardized DNA constructs, but its use of Type IIS restriction enzymes results in scarless assembly. Finally, the NEBuilder® HiFi DNA Assembly method from New England Biolabs® has minimal upfront requirements and allows you to expediently join multiple synthetic fragments, create multiple mutations in one or multiple fragments, and generate constructs for producing single-guide RNAs – but it also allows you to skip purification steps and end-repair steps with well-designed fragment overlap sections, while still creating scarless plasmid inserts.
Want to Learn More about DNA Assembly? Check Out Our Plasmids 101 eBook!
One of the features that makes NEBuilder HiFi so accurate (apart from the high-fidelity polymerase) is the ability to remove, reset and ligate mismatched overhangs, yielding scarless DNA assembly, and allowing fragments to be taken directly from restriction enzyme digests to DNA assembly. What makes it so forgiving, is the incorporation of a 3’-5’ exonuclease in the reaction mixture. The exonuclease removes mismatched 3’ overhangs, so that the region of homology doesn’t have to be located at the absolute end of a fragment. This means that fragments (containing regions of homology) cut from plasmids via restriction enzyme digestion, and used directly in DNA assembly will still yield scarless assemblies.
Overview of NEBuilder HiFi DNA assembly method
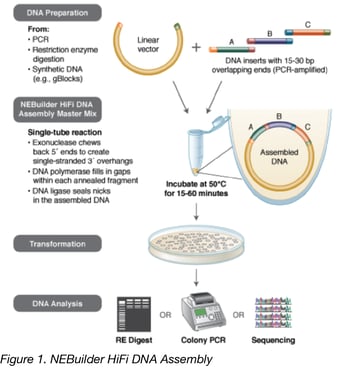 As with any well-executed cloning endeavor, NEBuilder requires upfront planning. This cloning methodology requires that the fragments to be joined contain regions of homology at, or close to, their ends. This homology can be contained within the individual fragments themselves, but doesn’t necessarily have to be. Regions of homology can be introduced to fragments via PCR using primers that contain the regions of homology. A powerful resource for planning an NEBuilder HiFi DNA Assembly mix reaction is the NEBuilder Assembly Tool which is freely available online. Its simple interface can guide you through both experimental and primer design based on fragment sequences and the polymerase used in the amplification. This tool allows instant updates as fragments are added, deleted, or reordered and PCR enzymes are adjusted. Appropriate fragment and primer design results in covalently joined, scarless DNA – perfect for transformation or as a PCR template.
As with any well-executed cloning endeavor, NEBuilder requires upfront planning. This cloning methodology requires that the fragments to be joined contain regions of homology at, or close to, their ends. This homology can be contained within the individual fragments themselves, but doesn’t necessarily have to be. Regions of homology can be introduced to fragments via PCR using primers that contain the regions of homology. A powerful resource for planning an NEBuilder HiFi DNA Assembly mix reaction is the NEBuilder Assembly Tool which is freely available online. Its simple interface can guide you through both experimental and primer design based on fragment sequences and the polymerase used in the amplification. This tool allows instant updates as fragments are added, deleted, or reordered and PCR enzymes are adjusted. Appropriate fragment and primer design results in covalently joined, scarless DNA – perfect for transformation or as a PCR template.
Some of the applications of the NEBuilder HiFi DNA Assembly Mix include:
- Generation of constructs with multiple point mutations
- Cloning sgRNAs
- Production of Yeast Expression Cassettes
Generating constructs with multiple point mutations
NEBuilder HiFi can simplify multi-position site-directed mutagenesis by assembling multiple fragments containing the desired mutations. This same process can be used simultaneously for the generation of one or more mutations – it is as easy as obtaining a plasmid with the gene of interest (search Addgene plasmids by gene here), designing primers with your desired mutation, performing PCR to generate the mutation and linearizing your vector (preferably by PCR) for DNA assembly. Further, there is no limit to where the mutation can be placed. With other techniques, when mutations are spaced farther apart, it becomes hard to accurately execute. However, these challenges can be circumvented by generating individual fragments containing mutations and aligned via regions of homology. With NEBuilder HiFi DNA assembly, it becomes as simple as mixing fragments together with linearized vector for seamless joining of multiple mutations spread over thousands of base pairs.
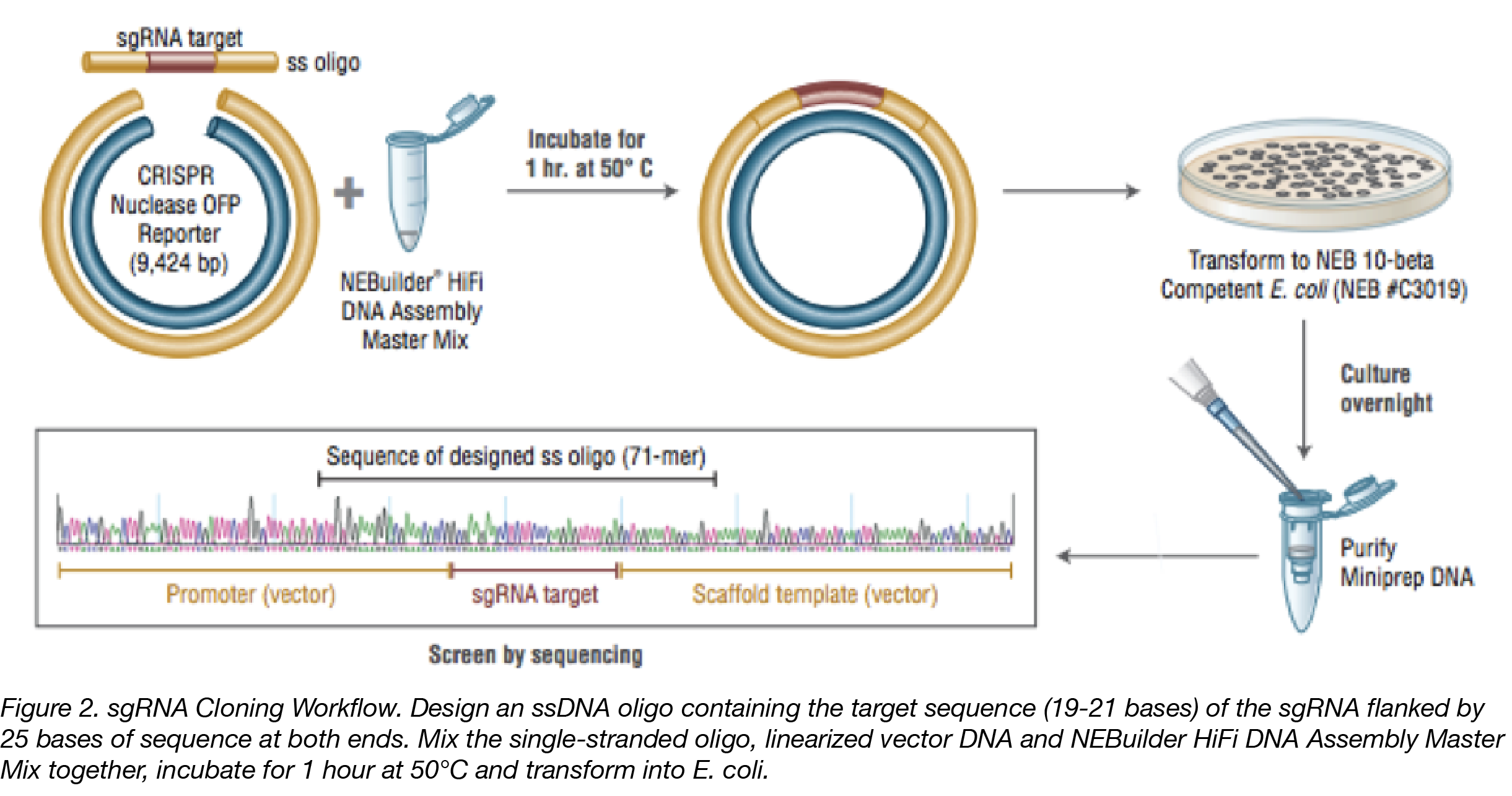
Cloning sgRNAs
Generating constructs to produce sgRNA for CRISPR experiments can be laborious, requiring multiple cloning steps. You can simplify this process by using NEBuilder which requires only a single-stranded synthetic oligo to generate a single-guide RNA (sgRNA). Using only one single-stranded oligo, instead of the more typical two oligos, is more cost effective and eliminates the annealing step usually used to create a double-stranded oligo. Generating an sgRNA construct becomes as simple as combining your favorite linearized plasmid with a synthetic oligo that contains a region of homology and any desired edits to the target region. Assembled plasmids can be directly transformed into E.coli for plating, screening and generation of sgRNA. Generation of sgRNA libraries, with maximum complexity, is achieved with synthetic oligos with a randomized region between the regions of homology.
Producing yeast expression cassettes
Traditionally, generating expression cassettes for yeast transformation can take 5 days. NEBuilder shortens that time to one day by simplifying the protocol to a single DNA assembly reaction. DNA fragments with the desired promoter, selectable marker, and target gene, for example, are amplified with overlapping regions of homology and inserted into a linearized cassette, ready for transformation into yeast that same day.
The field of synthetic biology has led to advances in biofuel generation, green chemistry and understanding the minimal genome. Many of these techniques require more complex DNA assemblies and improvements in these methodologies offer streamlined and simplified alternatives to traditional cloning. NEBuilder addresses the limitations associated with otherwise more complicated assembly approaches, and opens the door for new opportunities in the field.
Gibson Assembly® is a registered trademark of Synthetic Genomics, Inc.
One or more of these products are covered by one or more patents, trademarks and/or copyrights owned or controlled by New England Biolabs, Inc.
Many thanks to our guest blogger Lydia Morrison from New England Biolabs.
 Lydia Morrison is a biochemist by training and a content marketer by profession.
Lydia Morrison is a biochemist by training and a content marketer by profession.
Additional Resources on the Addgene Blog
- Learn More about Gateway Cloning
- Gibson Assembly and Other Long-Homology Based Cloning Methods
- Screen Plasmids by Colony PCR
Addtional Resources on Addgene.org
- Browse Our CRISPR Resources
- Find Validated gRNAs
- Browse All Plasmids
Topics: Plasmid Cloning, Plasmids






Leave a Comment