Regulating translation is key to cellular function, especially during development or stress. With ribosome profiling, researchers have been able to study the effects of various stimuli on global translation, but a visual technique to study translation remained elusive. Two techniques developed by Addgene depositors have made it easier to track translation in two different ways: by monitoring the first round of translation or by tracking the translation of a single mRNA over time. Both are helping researchers explore the complexity of translational control in cellular physiology.
Ribosome profiling
Ribosome profiling, a biochemical method developed in 2009, takes a “snapshot” of all of the mRNA bound-ribosomes in a cell. When cellular mRNAs are digested with ribonucleases, ribosomes protect the segments of mRNA to which they are bound. These regions are subsequently sequenced and aligned to the genome to determine genome-wide translational frequency. Although highly useful, this technique requires extensive sample processing to avoid disrupting ribosome-mRNA complexes. Recent work has adapted this technique to monitor local translation and determine if proteins are targeted to their destination co- or post-translationally.
TRICK: Visualizing the first round of translation
While ribosome profiling co-opts the physical location of the ribosome, TRICK (Translating RNA Imaging by Coat protein Knock-off) instead takes advantage of the ribosome’s movement. When the ribosome moves along an mRNA, it displaces other RNA-binding proteins to allow translation to occur. For a TRICK reporter mRNA this displacement leads to a change in the fluorescent signal: untranslated RNAs appear yellow, and translated RNAs appear red. To enable this color switching, TRICK requires the following components:
- a GFP protein that binds to the coding region of a reporter mRNA via a PP7 coat protein (NLS-PCP-GFP)
- an RFP protein that binds to the 3’ UTR of the reporter mRNA via an MS2 coat protein (NLS-MS2-RFP)
- the reporter mRNA, which contains the tethering sites for the fluorescent proteins and is driven by an inducible promoter (Plasmid 64542 or 64543)
Before translation, both GFP and RFP are bound to the reporter, causing it to appear yellow. The act of translation pushes NLS-PCP-GFP off of the mRNA, leaving only a red FP bound to the reporter. Because it contains a nuclear localization signal, NLS-PCP-GFP then returns to the nucleus to bind cognate untranslated mRNAs. The high specificity and resolution of this system are achieved by using multiple copies of the PP7 and MS2 coat protein binding sites to tether many copies of the FP-coat protein fusions to their reporter mRNAs. Using translation inhibitors cycloheximide and puromycin, Halstead et al. verified that translation is necessary for removal of GFP from the TRICK reporter.
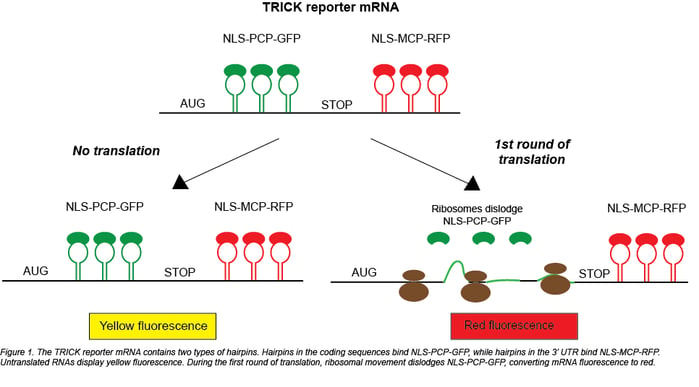 Delayed translation is known to be important in development, and Halstead et al. used TRICK to examine translation of Drosophila patterning gene oskar. Early in development, the osk-TRICK reporter was double labeled, indicating translational repression. During later stages, single RFP-labeled osk and Oskar protein were detected at the posterior pole. Oskar protein levels were negatively correlated with GFP intensity, showing that TRICK is an accurate readout of relative reporter translation.
Delayed translation is known to be important in development, and Halstead et al. used TRICK to examine translation of Drosophila patterning gene oskar. Early in development, the osk-TRICK reporter was double labeled, indicating translational repression. During later stages, single RFP-labeled osk and Oskar protein were detected at the posterior pole. Oskar protein levels were negatively correlated with GFP intensity, showing that TRICK is an accurate readout of relative reporter translation.
Single molecule continuous translation tracking
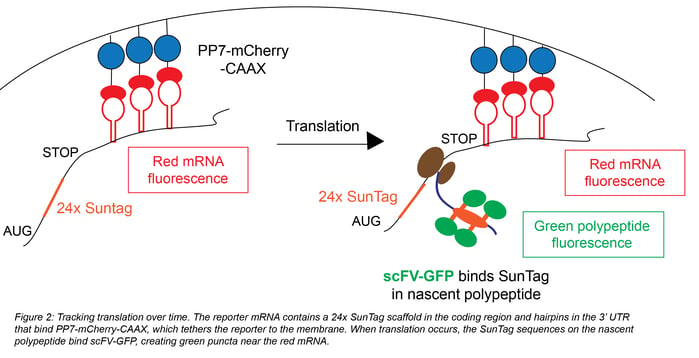
TRICK allows you to see the first round of translation, but it doesn’t allow continued tracking of a given mRNA because NLS-PCP-GFP is removed by the first ribosome that translates the mRNA. To permit tracking over time, Yan et al. devised a system that labels both mRNAs and their corresponding newly synthesized proteins. As in the TRICK system, the 3’ UTR of the reporter mRNA is labeled by PCP-mCherry (Figure 2). The 3’ UTR also contains a CAAX sequence to bind the mRNA to the plasma membrane; this sequence prevents diffusion of the mCherry labeled mRNA and keeps it in a single field of view for constant tracking. You can then directly monitor multiple rounds of translation from this single mRNA; you get a view of translational dynamics at the single molecule level.
The coding sequence of Yan et al.’s reporter contains 24 copies of SunTag, a synthetic scaffold that can recruit GFP fused to the SunTag-specific antibody scFv. Each time the reporter is translated, the SunTag sites on the nascent polypeptide recruit scFV-GFP, generating green puncta that colocalize with the red reporter mRNA. Yan et al. used harringtonine, a translation inhibitor that stalls ribosomes after initiation, to calculate a ribosome translocation rate of ~3.5 codons/second. They also explored the effects of 5’ UTR variants in translation of cell cycle protein Emi1, showing that ~80% of transcripts with a long 5’ UTR were translationally silent.
Both of these methods increase our understanding of translation dynamics, but they come with a few caveats. In both cases, creating a reporter mRNA requires inserting multiple hairpin sequences into the coding sequence and 3’ UTR. Before beginning experiments, it’s important to verify that these changes do not impact translation rate, as the authors of these papers did. Since the SunTag-based system tethers mRNAs to the plasma membrane, it’s not appropriate for use with mRNAs usually translated in a specific compartment. Despite these limitations, these techniques represent powerful tools for studying translation in various cell types and physiological states.
References
1. Ingolia, Nicholas T., et al. “Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling.” Science 324(5924) (2009): 218-23. PMID: 19213877 PMCID: PMC2746483
- Find ribosome profiling plasmids at Addgene.
2. Halstead, James M., et al. “Translation. An RNA biosensor for imaging the first round of translation from single cells to living animals.” Science 347(6228) (2015): 1367-71. PMID: 25792328 PMCID: PMC4451088
- Find plasmids from this paper at Addgene.
3. Yan, Xiaowei, et al. “Dynamics of Translation of Single mRNA Molecules In Vivo.” Cell 165(4) (2016): 976-89. PMID: 27153498 PMCID: PMC4889334
- Find plasmids from this paper at Addgene.
Additional Resources the Addgene Blog
- Learn More about SunTag
- Catch up on Your GFP Background
- Choose the Brightest Fluorescent Protein
Resources on Addgene.org
- Find Fluorescent Protein Plasmids
- Check Out Other Hot Plasmid Technologies
Topics: Fluorescent Proteins, Localization with Fluorescent Proteins






Leave a Comment