In biology as in life, more is often better. More transcription factor binding sites in a promoter lead to higher transcriptional activation. Multiple nuclear localization signals (NLS) increase protein import into the nucleus. In developing their SunTag technology, the Vale and Weissman labs took this biological lesson and created a system to amplify fluorescent signals. Named for the "stellar explosion SUperNova," SunTag can help you turn up the brightness in your fluorescent imaging experiments.

Fluorescent protein fusions
One of the easiest ways to track a given protein is to fuse it to a fluorescent protein. You can then study where the protein is localized, and how its localization and expression may change across various conditions. However, this system is far from perfect. For example, If your fusion protein is expressed at low levels, you have to increase your imaging time to get enough signal. This workaround risks cellular phototoxicity and eliminates the possibility of long-term imaging studies. If you instead overexpress your protein at higher levels, you risk observing artifacts only present when the protein is at a very high concentration. Overexpressed proteins also have the potential to form aggregates and may be toxic to the cell.
Here comes the SunTag
How does SunTag fix these problems? Instead of directly fusing a fluorescent protein to your protein of interest, you instead fuse it to the synthetic SunTag scaffold. This scaffold contains 10-24 copies of the short epitope GCN4. GCN4 recruits GFP fused to the cognate scFV antibody, which is expressed from a separate plasmid. This system amplifies the intensity of the fluorescent signal and enables single molecule tracking within living cells without affecting protein function, thereby creating a single-molecule reporter of intracellular processes. Initially, Tanenbaum et al. observed some GFP aggregation, which they reduced by using superfolder GFP (sfGFP) with the small solubility tag GB1. In SunTag nomenclature throughout the rest of this blog post, GFP refers to sfGFP-GB1. 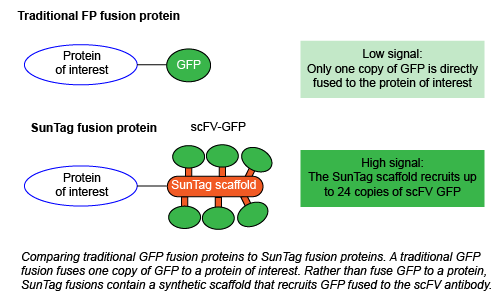
Tanenbaum et al. examined the power of SunTag for single molecule imaging, finding that plasma membrane-targeted CAAX-SunTag was 18-fold brighter than sfGFP! The high SunTag signal allowed them to cut their laser power by over 80% and still obtain a higher signal-to-noise ratio with a lower photobleaching rate. Given the power of SunTag, they attempted single-molecule imaging deep inside the cell, in the nucleus and cytoplasm. Again, they found that SunTag marked single molecules very effectively - they even managed to track run lengths of the motor protein kinesin across microtubules.
Tanenbaum et al. then tested their hypothesis that the lower expression levels of SunTag constructs would avoid negative effects on cell physiology. Having seen that mitochondrial tracker GFP-mitoNEET can impair mitochondrial function, they examined the effects of mitoNEET-SunTag-GFP. As expected, they obtained bright images of mitochondria without organelle toxicity.
First generation (v1) SunTag is expressed at very low levels due to poor stability of the GCN4 scaffold. To increase expression, Tanenbaum et al. modified the GCN4 sequence to increase its alpha-helical structure and stability, creating v4 SunTag. Since the v4 system does not display protein aggregation, it’s recommended for most imaging applications.
What can you do with SunTag?
Tanenbaum et al. performed a variety of different experiments in their paper, and so can you! In almost any case you would use a traditional FP fusion, a SunTag fusion could be used as well.
Pros of SunTag compared to traditional fluorescent proteins
- Improved brightness and signal-to-noise ratio
- Less chance of phototoxicity
- Reduced photobleaching
- Simplified, long-term single-molecule tracking
Caveats of SunTag
- Very large size: a 24x scaffold fully occupied with GFP has a molecular weight of 1,400 kDa vs 24 kDa for GFP alone. Although traditional FP fusions can also affect protein activity, half-life, or localization, these concerns are greater with SunTag.
- v1 SunTag exhibits some scaffold aggregation (v4 Suntag does not.)
It’s up to you to determine SunTag’s suitability in your experiments on a protein-by-protein basis. However, since many of Addgene’s SunTag plasmids have coveted “blue flames,” it’s clear that the system is broadly applicable across many areas of research. Beyond fluorescence, SunTag can also be used to improve CRISPR-based activation of target genes, but we’ll save that application for another day!
References
1. Tanenbaum, Marvin E., et al. “A protein-tagging system for signal amplification in gene expression and fluorescence imaging.” Cell 159(3) (2014):635-46. PubMed PMID: 25307933. PubMed Central PMCID: PMC4252608.
Additional Resources on the Addgene Blog
- Sign Up to Receive the Fluorescent Protein eBook
- Avoid Fluorescent Protein Oligomerization
- Learn about Small Molecule Fluorphores
Resources on Addgene.org
- Find Fluorescent Protein Empy Backbones
- Visit Our Fluorescent Protein Guide Page
- Find Luciferase Plasmids
Topics: Fluorescent Proteins, Generating Fusions





Leave a Comment