One of the main tenets of our quality control (QC) mantra is to do our utmost to ensure that scientists are receiving the exact materials that they think they are. To address this, several years ago we partnered with Seqwell to establish a next-generation sequencing (NGS) QC platform that allows us to provide scientists with complete plasmid sequences. After launching our viral service in 2016, we adapted this platform to accommodate adeno-associated viral vector (AAV) samples and created a simplified process, termed viral genome sequencing (VGS), to confirm the identity and serotype of our AAV preparations. We recently published a description of the VGS process and use cases in Human Gene Therapy.
Existing methods for AAV quality control
Historically, AAV vector QC has been sparse including only titration of AAV particles and a protein stain to assess purity. For labs that produce a small number of AAV preparations this level of quality control is sufficient but for viral vector core facilities producing a large number of viral preparations in parallel, there is a very real risk of mixing up samples, serotypes, or both. Unfortunately, the standard AAV QC regimen does not address this.
Previous studies have demonstrated that NGS is an effective tool to confirm AAV vector genome identity. These studies used SMRT sequencing, TA-based ligation and tagmentation-based methods for AAV genome sequencing (Tai 2019; Lecomte 2015; Maynard 2019). Unfortunately, at present SMRT sequencing only works for self-complementary AAVs and the TA-based ligation and tagmentation preparation approaches described previously require a preliminary double stranding step. The double stranding step was assumed to be necessary because the AAV genome is single-stranded and both TA-based ligation and tagmentation require a double-stranded template.
Using viral genome sequencing to verify the viral genome
We hypothesized that this double stranding step might not actually be necessary. Since the AAV genome exists as [+] and [-] strands that are packaged at the same frequency, we reasoned that, following DNA extraction, the [+] and [-] strands might naturally associate forming a double-stranded species that could be used as a template for downstream applications (Berns, 1972). Using qPCR and restriction enzyme based methods, we showed that the DNA extract is a heterogeneous mix of single-stranded and double-stranded species. While not all of the molecules in the DNA extract associate with complementary strands, enough double-stranded species exist in the mix to serve as effective substrates for tagmentation-based library preparation procedures. This finding eliminates the double-stranding step and reduces both the time and cost for AAV genome sequencing.
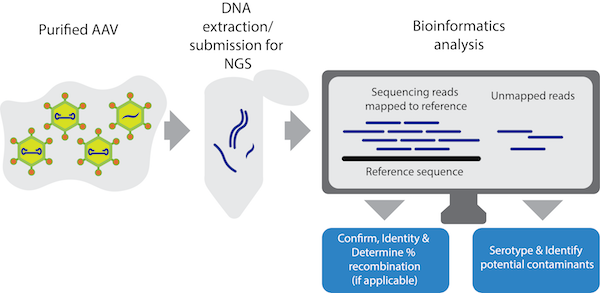 |
| Figure 1: The viral genome sequencing workflow. |
Using this simplified approach, we sequenced over 100 AAV vector preparations and showed that VGS can quickly and reliably confirm the identity of AAV genomes and detect contaminating AAV DNA. In addition, we wrote an open-source custom Python script that uses the VGS data to determine the percent of recombination present in Cre-dependent vectors and a script that can confirm the serotype of AAV preparations.
Using viral genome sequencing to determine serotype
AAV packaging is not perfect and while we think of the AAV vector genomes as containing just the expression cassette that we want, it also includes a small amount of spurious DNA from the packaging cell line, helper and packaging plasmids. We decided to use this to our advantage by designing a Python script that looks for pre-defined serotype specific signature sequences in the data. Unlike previously described thermodynamics-based methods of serotype confirmation, this method does not require any additional reagents or lab work (Pacouret, 2017). Moreover, the sequencing data from multiple AAV lots can be run in parallel and provide serotype data in minutes and the scripts can be easily customized to detect other sequences of interest.
While the VGS process isn’t new to Addgene, we are excited to share the procedures and Python scripts with laboratories and viral vector cores routinely producing AAV. We believe that this simplified sequencing and analysis approach can be easily incorporated into standard QC regimens and will provide scientists with the much needed assurance that their research materials are exactly what they think they are.
References
Berns KI, Adler S (1972) Separation of two types of adeno-associated virus particles containing complementary polynucleotide chains. J Virol 9(2): 394-396.
Lecomte E, Tournaire B, Cogné B, Dupont J-B, Lindenbaum P, Martin-Fontaine M, Broucque F, Robin C, Hebben M, Merten O-W, Blouin V, François A, Redon R, Moullier P, Léger A (2015) Advanced Characterization of DNA Molecules in rAAV Vector Preparations by Single-stranded Virus Next-generation Sequencing. Molecular Therapy - Nucleic Acids 4:e260 . https://doi.org/10.1038/mtna.2015.32
Maynard LH, Smith O, Tilmans NP, Tham E, Hosseinzadeh S, Tan W, Leenay R, May AP, Paulk NK (2019) Fast-Seq: A Simple Method for Rapid and Inexpensive Validation of Packaged Single-Stranded Adeno-Associated Viral Genomes in Academic Settings. Human Gene Therapy Methods 30:195–205 . https://doi.org/10.1089/hgtb.2019.110
Pacouret S, Bouzelha M, Shelke R, Andres-Mateos E, Xiao R, Maurer A, Mevel M, Turunen H, Barungi T, Penaud-Budloo M, Broucque F, Blouin V, Moullier P, Ayuso E, Vandenberghe LH (2017) AAV-ID: A Rapid and Robust Assay for Batch-to-Batch Consistency Evaluation of AAV Preparations. Molecular Therapy 25:1375–1386 . https://doi.org/10.1016/j.ymthe.2017.04.001
Additional resources on the Addgene blog
- Read all of our viral vectors related blog posts
- Read our blog post on AAV titers
- Browse viral vectors protocols and tips
Resources on Addgene.org
- Find AAVs at Addgene
- Learn more about Addgene's viral service
- Find protocols for working with virus
Topics: Viral Vectors, Viral Vector Protocols and Tips, AAV






Leave a Comment