This post was contributed by guest blogger Natalie Niemi, a postdoctoral fellow at the Morgridge Institute for Research in Madison, Wisconsin.
 It is commonly cited that approximately one-third of cellular proteins are modified through phosphorylation (1). However, the expansion of studies on protein phosphorylation in an array of model systems coupled with advances in mass spectrometry suggest that phosphorylation is far more prevalent than previously appreciated. PhosphoSitePlus, one of the most inclusive databases of post-translational modifications, identifies a staggering ~250,000 phosphorylation events in the proteomes of higher mammals (2). How can we begin to understand the importance of any of these phosphorylation events on the activity of a given protein?
It is commonly cited that approximately one-third of cellular proteins are modified through phosphorylation (1). However, the expansion of studies on protein phosphorylation in an array of model systems coupled with advances in mass spectrometry suggest that phosphorylation is far more prevalent than previously appreciated. PhosphoSitePlus, one of the most inclusive databases of post-translational modifications, identifies a staggering ~250,000 phosphorylation events in the proteomes of higher mammals (2). How can we begin to understand the importance of any of these phosphorylation events on the activity of a given protein?
Classical methods for studying protein phosphorylation
A number of classic methods exist for this, the most common of which is the utilization of phosphomimetic residues. The phosphorylated residue of interest is mutated to a negatively charged residue, aspartate or glutamate, imparting the negative charge associated with phosphorylation. However, these amino acids fail to recapitulate the true steric and charge-based nature of the phosphoryl-modification; they are both much smaller, and carry only a single negative charge as opposed to a double negative charge in the phosphate group. Additionally, these mutations are not reversible, and thus cannot reflect the true state of protein modification within the context of its appropriate regulation.
To avoid these problems, it is also possible to isolate a phosphorylated intermediate of a protein for characterization. However, phosphoproteins are often low abundance, and thus difficult to purify in quantities necessary for study. Moreover, the ability to isolate purified phosphorylated products usually depends on tools such as phosphospecific antibodies, which aren’t available for the majority of documented phosphorylation sites. If knowledge of a candidate regulatory kinase is known, it’s possible to generate non-phosphorylated recombinant protein and perform in vitro kinase reactions. Often, however, researchers lack knowledge of the regulatory kinase, access to active kinase for the reaction, or have difficulties with kinase specificity in vitro. The establishment of a system in which purified, recombinant protein could be produced with site-specific incorporation of phosphoserine (as described below) provides researchers with a valuable tool for understanding the contribution of phosphorylation to enzyme function.
The development of phosphoserine incorporation
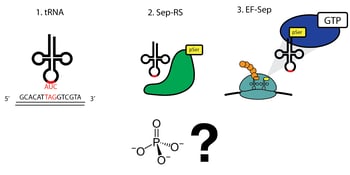
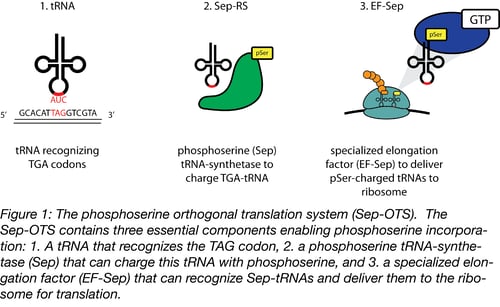
Extensive genomic sequencing of prokaryotic lineages revealed a curious anomaly amongst a subset of methanogenic archea: these organisms lacked a classic cysteinyl tRNA synthetase, despite abundant incorporation of cysteine in their proteomes (3). This led to the eventual discovery of a naturally occurring phosphoserine-tRNA synthetase that serves as an intermediate on the path to the formation of cysteinyl-tRNA (4). The discovery of a phosphoserine (Sep)-tRNA synthetase enabled researchers to create an orthogonal translation system (OTS) for the incorporation of phosphoserine directly into recombinant proteins (5). Optimization of this system defined three components required for translation (Figure 1):
- A tRNA that recognizes a specific ‘open’ codon and utilizes this triplet to incorporate phosphoserine
- A Sep-tRNA synthetase, which charges this tRNA with phosphoserine
- A specialized elongation factor that allows proper extension and translation of the pSer-incorporated polypeptide.
These three components were shown to effectively incorporate phosphoserine into recombinant proteins, such as the kinase MEK1 (5). MEK1 is inactive without requisite dual phosphorylation events on its activation loop, but can be made constitutively active through phosphomimetic mutation of these residues. Due to the limitations previously mentioned, phosphomimetic mutations on the activation loop do not fully activate MEK1. In fact, replacing just one of the MEK1 phosphomimetic residues in the above study with phosphoserine (to create a pSer/phosphomimetic dual mutant) elevated kinase activity over 70-fold over dual phosphomimetic mutation. This emphasizes the advantages of bona fide pSer-incorporation into recombinant proteins as an alternative to phosphomimetic mutation.
Creating phosphoserine-incorporated proteins
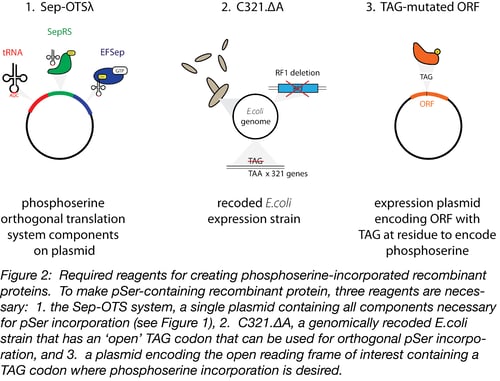
The generation of pSer-incorporated proteins requires a molecular toolkit containing the OTS, an appropriate bacterial host strain for protein production, and an expression plasmid encoding the open reading frame (ORF) of interest (Figure 2).
1. The phosphoserine OTS
The backbone of the phosphoserine OTS, Sep-OTS, contains the components required for the translation of pSer-incorporated proteins on a single plasmid. This includes 4 copies of a tRNA that specifically recognizes amber stop codons (TGA) to encode phosphoserine, the Sep-tRNA synthetase, and EF-Sep – an optimized elongation factor that accepts Sep-charged tRNAs during translation. Many ‘flavors’ of Sep-OTS plasmids are available through Addgene, but we have worked most extensively worked with and optimized Sep-OTSλ and can recommend its use (6).
2. C321, a specialized bacterial host strain for pSer-incorporation
The second component necessary for phosphoserine incorporation is a modified bacterial strain, C321.ΔA. Because the phosphorserine OTS recognizes the TAG amber stop codon, using the OTS in an unmodified bacterial strain would result in read-through translation of many stop codons and detrimental effects on cell growth. The C321.ΔA strain was genomically recoded to eliminate all 321 amber stop codons (TAG), replacing them with TAA (7). This modification allowed the deletion of release factor 1, the protein responsible for terminating translation in response to the presence of TAG sequences in ORFs. In this bacterial strain, TAG is an ‘open’ codon, which can be specifically recognized by the Sep-tRNA.
3. An expression plasmid encoding the ORF to be expressed
Finally, the researcher needs a plasmid encoding their open reading frame of interest. Two expression systems have been verified for use by the Rinehart lab: pCRT7 NT TOPO E17TAG GFP and pGEX-6P-1. The pCRT7 NT TOPO vector contains a mutant pSer-containing GFP that can be used as a positive control during optimization experiments. This version of GFP will contain a phosphoserine at amino acid 17 instead of a glutamate which allows the system of interest to be validated first through the production of full-length protein (the TAG codon should encode phosphoserine instead of a stop codon, which will allow full length GFP to be translated), and second, through a gel shift assay using the PhosTag system (see below). This plasmid backbone can also be cloned with an ORF of interest for customized experiments. Once the ORF of interest is cloned into one of these expression vectors, the codon for the amino acid to be replaced with phosphoserine must be recoded to ‘TGA’ using site-directed mutagenesis. In the C321.ΔA strain, this TGA will be recognized by Sep-tRNA and translated with a specific phosphoserine residue at this position.
Practical tips for your experiments
To generate as much pSer-incorporated protein of interest as possible, some optimization will be necessary. To begin, one should optimize protein production using standard protein purification strategies; expression should be optimized for parameters such as induction temperature and length of induction, which will vary between different proteins. Beyond these standard considerations, it is important to determine how much phosphorserine has actually been incorporated into the full-length protein. Up to 80% p-Ser incorporation into E17TAG GFP has been achieved with the Rinehart system (6); however different proteins will likely achieve different levels of incorporation due to differences in stability of the p-Ser incorporated protein as well as context-specific effects of UAG suppression, as described (6). To determine the stoichiometry of phospho-incorporated to non-phospho-incorporated proteins purified from the system, researchers can run samples on SDS-PAGE gels containing Phos-Tag, a small molecule that binds specifically to phosphate moieties and promotes a robust mobility shift in phosphorylated, but not non-phosphorylated proteins (8). Quantifying the ratio of phospho- and non-phospho-bands will give the researcher an idea of the purity of pSer-incorporated to non-phosphorylated protein. Verification of site-specific pSer incorporation can also done using mass spectrometry to identify a mass shift proportional to a single phosphate moiety on the peptide of interest.
After confirming pSer incorporation, researchers can demonstrate effects of pSer on enzyme function, protein interaction, etc. by comparison to non-phosphorylated or ‘wild type’ control protein. The ‘wild type’ control can be made one of two ways. First, co-expression of a suppressor tRNA (SupD) instead of the SepOTSl can encode a non-phosphorylated serine at the TAG codon of the same expression vector used for phosphoserine incorporation (6). This will give the researcher both a phosphoserine and ‘wild type’ (with the exception of the incorporation of serine) population of protein using the same expression plasmid. This is a particularly important control when creating a phosphoserine-incorporation mutant for a residue that wasn’t originally a serine (e.g. mutating a threonine residue to a phosphoserine) to control for effects of the amino acid substitution. Alternatively, you can express the completely wild type protein from the same plasmid.
Upon verifying that serine phosphorylation affects protein function, researchers should try to reverse the effects of pSer-incorporation on enzyme function through phosphatase treatment. If a candidate phosphatase is unknown, CIP or other generic phosphatases may dephosphorylate the pSer-incorporated protein, ideally bringing the phenotype assayed closer to that of the wild type protein. The efficiency of the dephosphorylation reaction can be tracked through the comparison of phospho:non-phospho species on PhosTag gels as described above.
Reagents and protocols
Sep-OTSλ plasmid - Addgene # 68292
C321.ΔA bacterial strain – Addgene #68306
pCRT7 NT TOPO E17TAG GFP plasmid - Addgene #68295
SupD plasmid – Addgene # 68307
For further technical reference and protocols for use of this system, please reference ‘Rinehart Lab reagents for improved expression of recombinant phosphoproteins’ available on the Addgene website.
Many thanks to guest blogger Natalie Niemi!
 Natalie Niemi, Ph.D., is a postdoctoral fellow at the Morgridge Institute for Research in Madison, Wisconsin. She is particularly interested in the intersection of cell signaling, metabolism, and the mighty mitochondrion. You can follow her on Twitter @nieminm.
Natalie Niemi, Ph.D., is a postdoctoral fellow at the Morgridge Institute for Research in Madison, Wisconsin. She is particularly interested in the intersection of cell signaling, metabolism, and the mighty mitochondrion. You can follow her on Twitter @nieminm.
References
1. Cohen, Philip. "The origins of protein phosphorylation." Nature cell biology4.5 (2002): E127-E130. PubMed PMID: 11988757.
2. Hornbeck, Peter V., et al. "PhosphoSitePlus, 2014: mutations, PTMs and recalibrations." Nucleic acids research 43.D1 (2015): D512-D520. PubMed PMID: 25514926. PubMed Central PMCID: PMC4383998.
3. Li, Tong, et al. "Cysteinyl‐tRNA formation: the last puzzle of aminoacyl‐tRNA synthesis." FEBS letters 462.3 (1999): 302-306. PubMed PMID: 10622715.
4. Sauerwald, Anselm, et al. "RNA-dependent cysteine biosynthesis in archaea." Science 307.5717 (2005): 1969-1972. PubMed PMID: 15790858.
5. Park, Hee-Sung, et al. "Expanding the genetic code of Escherichia coli with phosphoserine." Science 333.6046 (2011): 1151-1154. PubMed PMID: 21868676.
6. Pirman, Natasha L., et al. "A flexible codon in genomically recoded Escherichia coli permits programmable protein phosphorylation." Nature communications 6 (2015). PubMed PMID: 26350500. PubMed Central PMCID: PMC4566969.
7. Lajoie, Marc J., et al. "Genomically recoded organisms expand biological functions." Science 342.6156 (2013): 357-360. PubMed PMID: 24136966.
8. Tomida, J., et al. "Detection of phosphorylation on large proteins by Western blotting using Phos-tag containing gel." Nat. Protoc. doi 10 (2008). Journal.
Resources on the Addgene Blog
- Verify Your Constructs with Colony PCR
- Read about Tags for Protein Purification
Resources on Addgene.org
Topics: Synthetic Biology, Other Plasmid Tools, Plasmids






Leave a Comment