Much of today's biological research requires a close examination of specific proteins within a system. This can be pretty complicated given that a single cell has tens of thousands of proteins functioning in a variety of ways. How do scientists focus on the activity or function of a single protein? Oftentimes, they rely on protein purification. In this strategy a single protein can be isolated from a complex mix using affinity chromatography against an amino acid sequence, or tag, on the protein. While a variety of methods can be used to successfully isolate proteins, this blog will focus on one of the most common, purification based on the polyhistidine tag.
What is the polyhistidine tag?
The polyhistidine tag is a string of 6 or more histidine residues expressed in frame on the N- or C-terminus of a recombinant protein of interest. The tag was developed as a means to purify a protein based on the affinity of the imidazole ring of histidine for metal ions such as Ni2+ or Co2+ (Hochuli 1988), and can elute proteins with up to 95% purity (Janknecht 1991, Hochuli 1988).
In this purification approach, the metal ions are immobilized on a bead or resin and a sample containing the protein of interest flowed through. The protein of interest binds to the metal ions, becoming immobilized itself. Next, a series of wash steps removes excess proteins and other contaminants. The protein of interest is then eluted using a high imidazole buffer that outcompetes the interaction between the protein of interest and the metal ions.
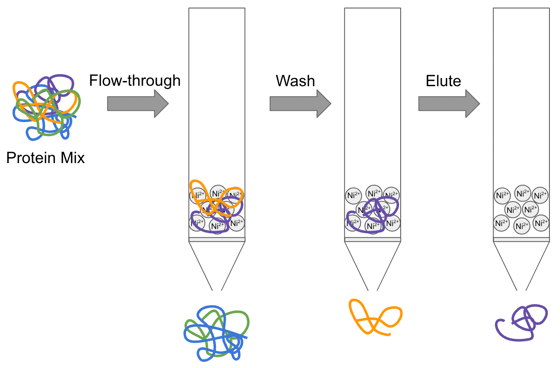
|
| Fig. 1: A sample containing a polyhistidine tagged protein of interest (purple) and several contaminating proteins (blue, green, orange) flows through the Ni2+-containing resin. The protein of interest (purple) binds to the Ni2+ and is immobilized along with a polyhistidine containing contaminating protein (orange). A low imidazole wash buffer disrupts the weak interaction of the contaminating protein but not that of the protein of interest. A high imidazole elution buffer disrupts the interaction of the protein of interest and it is successfully isolated. |
Prior to the polyhistidine tag, alternative fusion tags were used, such as S. aureus’ Protein A (Moks 1987). These tags, however, tended to be very large and often required the incorporation of a cleavage site and a post-purification cleavage step to remove the tag in order to prevent it interfering with protein function. In contrast, the polyhistidine tag is very small and rarely affects protein function (Bornhorst 2000).
One downside of polyhistidine tag-based purification, however, is the propensity for nonspecific binding of untagged proteins. Despite being a relatively low frequency amino acid, proteins containing two or more adjacent histidines are present in most production systems and can bind and co-elute with the protein of interest (Bornhorst 2000). To prevent this, wash steps often include low concentrations of imidazole. The concentration of imidazole used in the wash buffer should be high enough to disrupt contaminant-metal ion binding without eluting the target protein and needs to be empirically determined. In addition, some metal ion substrates, such as those containing Co2+, have a lower affinity for polyhistidine than more commonly used substrates such as Ni2+ and are therefore more selective. When using systems with higher levels of histidine-containing protein contamination, such as mammalian production systems, consider using these more selective substrates along with stringent wash conditions.
Find polyhistidine tag plasmids here!
Where should I place the tag and how many do I need?
When designing a plasmid containing a polyhistidine tag, you’ll need to decide whether to add the tag to the N- or C-terminus and how many histidine residues to include.
Placement
As far as placement goes, there is no cut or dried answer. Successful purification requires the tag to be accessible to the metal ions once the protein is folded. Sometimes a tag placed on one end will be blocked during protein folding, in which case you should try tagging the opposite end.
If neither placement works, consider purifying under denaturing conditions. Unlike other affinity purification methods where binding relies on a specific protein conformation, the polyhistidine-metal ion interaction is not disrupted by denaturants (Hochuli, 1988). In some cases, proteins can be bound under denaturing conditions to expose the tag and then refolded by gradual removal of the denaturant. Not all proteins will refold once denatured, however, so take care when using this method.

|
| Fig 2: A, The polyhistidine tag is a string of histidine amino acids on the N- or C- terminus of a protein. Neither the N or C terminus is preferable over the other however, tag placement can affect protein function. B, The string of histidines in the tag is typically between 6 to 9 residues long. The small size of the polyhistidine tag minimizes the chance of the tag disrupting normal protein function, however longer histidine strings can increase that risk. |
Number of residues
Deciding how many histidine residues to use is a bit easier. The more histidine residues that are present, the more tightly the protein will bind to the metal ion substrate. If your protein of interest is expressed at a very low level, try a longer histidine tag to increase capture. Longer histidine tags are also recommended if you need a very pure sample. Because binding strength is proportional to the length of the tag, with longer tags you can use more stringent wash conditions and remove more contaminants without losing your protein of interest. But that doesn’t mean longer is always better - the longer the tag, the bigger it is, and the greater the risk that it will interfere with protein function.
Additional Benefits of HIS tags
In addition to providing a way to purify a protein of interest, polyhisitidine tags can also be used as a target for antibody based assays. Antibodies are useful tools for studying proteins and are used for a variety of assays in the lab (for a refresher see Antibodies 101: Introduction to Antibodies.) Unfortunately, for some proteins there just aren’t any good antibodies available. In these cases, having a tagged version of the protein is hugely beneficial to scientists. If their protein of interest is fused to polyhistidine, then they can use an anti-HIS antibody, like this one developed by Dr. James Trimmer’s lab, to perform assays that they otherwise would not be able to do.
We hope that this blog has answered all of your questions about polyhistidine tags. For more antibody-related information, sign up to receive alerts for antibody posts from our blog!
Resources and References
Resources on addgene.org
Ready-to-use antibodies from Addgene
More Resources on the Addgene blog
Antibodies 101: Introduction to Antibodies
References
Hochuli E, Bannwarth W, Döbeli H, Gentz R and Stüber D. Genetic Approach to Facilitate Purification of Recombinant Proteins with a Novel Metal Chelate Adsorbent. Bio/Technology. 1988; 6: 1321–1325.
Janknecht R, de Martynoff G, Lou J, Hipskind RA, Nordheim A, Stunnenberg HG. Rapid and efficient purification of native histidine-tagged protein expressed by recombinant vaccinia virus. Proc Natl Acad Sci U S A. 1991;88(20):8972-6.
Moks T, Abrahrnsen L, Oesterlof B, Josephson S, Oestling M, Enfors SO, Persson I, Nilsson B and Uhlen M. Large-scale affinity purification of human insulin-like growth factor I from culture medium of E.coli. Bio/Technology. 1987, 5:379-382.
Bornhorst JA and Falke JJ. Purification of Proteins Using Polyhistidine Affinity Tags. Methods Enzymol. 2000; 326: 245–254
Topics: Antibodies






Leave a Comment