You may have heard the term antibody tossed around in the news or in the lab. But what exactly is an antibody, and how is a component of the immune system useful as a research reagent? Let’s find out!
What is an antibody?
Antibodies, also known as immunoglobulins, are ~150 kDa, Y-shaped proteins that are both a natural part of the immune system and a tool that can be used for a variety of research applications. Within the immune system, antibodies are produced by B cells. They bind to proteins on the surface of extracellular pathogens such as parasites or microbes, or to proteins expressed on the surface of cells that have been infected with a microbe, to trigger immune cascades that clear these infections. Anything that generates an antibody response in the immune system is referred to as an antigen. You can learn more about antibodies in our Antibodies 101 animation, What is an Antibody?
The ability of antibodies to bind proteins is useful for research applications as well because they allow scientists to target specific proteins they’re interested in. Once a protein is targeted with an antibody, you can visualize the protein via fluorescence or chemiluminescence, precipitate the protein out of solution, or isolate cells expressing this protein. Read on to learn more about antibodies and how to use them in the lab!
Parts of an antibody
Antibodies have two regions: the Fab, or antigen-binding region, and the Fc, or crystallizable region. Two immunoglobulin (Ig) heavy chains and two Ig light chains make up each antibody molecule. The Fc region and part of the Fab regions are made from the Ig heavy chains, and the rest of the Fab regions are completed with the Ig light chains. The variable region of the heavy and light chain determine what antigen the antibody recognizes and binds to.
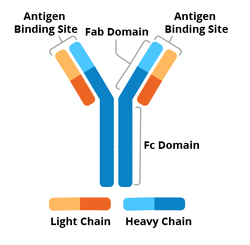 |
| Figure 1: Labeled diagram of an antibody including Fc, Fab, heavy chain, light chain, constant region, variable region |
The constant region of the heavy chain determines the antibody’s isotype - that is, one of five broad groups of antibodies (IgM, IgD, IgA, IgG, or IgE). In the context of the immune system, each isotype is expressed at different times of the immune response and initiates different immune cascades. In the research setting, antibodies of different isotypes can be used together in the same experiment because the reagents used to detect the presence of an antibody depends partially on its isotype.
The variable region of an antibody does not recognize the entirety of the target protein: instead, it recognizes a small portion of the protein, known as an epitope. Epitopes are between 5-8 amino acids long, with a typical length of 5 or 6 amino acids (Cruse et al., 2004). Because proteins are much larger than this, it may contain many epitopes that antibodies can recognize. Imagine you’re doing a word search puzzle. You’re not always going to spot the whole, long word right off the bat, but you might see a few letters and be able to guess whether or not that string of letters is a part of the word you’re searching for. Antibodies may recognize a linearized sequence of amino acids, or only in their 3D conformation.
Types of antibodies and production
In the immune system, antibodies are produced by B cells. But, there are actually many ways that antibodies can be produced for research purposes, some of which take advantage of natural immune system production and many more engineered methods. Each type of antibody and production strategy has advantages and disadvantages compared to each other.
Polyclonal antibodies
These antibodies are derived from animals, such as rabbits or goats; the animals are injected with the antigen that you want the antibody to recognize. This triggers an immune response within the animal, and its B cells will churn out antibodies against the injected protein. Several weeks later, blood from the animal is drawn and the antibodies are extracted. These antibodies recognize a variety of epitopes on the target protein, so they are referred to as polyclonal. Polyclonal antibodies have the advantage of more robustly recognizing the target protein because the antibodies will recognize multiple epitopes on the protein. However, each animal will make antibodies against different protein epitopes and so variability between lots is a big concern. You can find more details about polyclonal antibodies on our Antibodies 101: Polyclonal Antibodies blog post.
Monoclonal antibodies
On the other hand, monoclonal antibodies are a homogenous population of antibodies that all recognize the same epitope of the target protein. Most commonly these antibodies are produced from hybridomas. Hybridomas are formed by fusing B cells that have been extracted from animals injected with protein antigen to myeloma cells. The hybridomas are then subcloned by limiting dilution, which produces an immortalized, clonal population of cells that produce a single variant of antibody. Monoclonal antibodies are less likely to cross-react with other, similar proteins due to recognizing a single epitope of the target protein. However, small mutations in the DNA of the hybridoma can occur, known as genetic drift, meaning that over time, the antibody you expect it to produce is not necessarily what you are getting. Learn more about monoclonal antibodies here.
Recombinant antibodies
Similar to monoclonal antibodies, recombinant antibodies are a homogeneous population. However, unlike monoclonal antibodies, which come from naturally-occurring immunoglobulin genes, recombinant antibodies are plasmid-based. Their sequences can be optimized for specificity and sensitivity, for example by changing certain amino acids to improve its stability. Additionally, the choice of vector can impact the expression level of an antibody (Ayyar et al., 2017).
Single-chain variable fragment (ScFv)
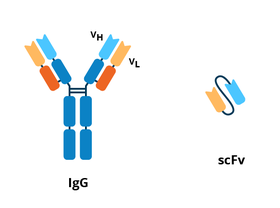 |
| Figure 2: Comparison between the IgG antibody and scFv. |
Because the entire antibody molecule is not necessary for antigen binding, the variable regions alone can be generated as a fusion protein. An ScFv is made up of the variable regions of the heavy and light chains fused together to form a single protein that can recognize the target protein (Wang et al., 2013). These proteins are also plasmid-derived. However, a disadvantage to ScFv is their lower affinity for their target protein compared to whole antibodies (Crivianu-Gaita 2016). Get more details on ScFvs in this article: Antibodies 101: Single Chain Fragment Variables (scFvs).
Nanobodies
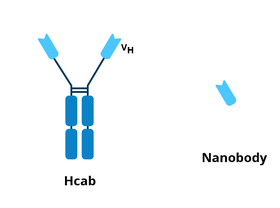 |
| Figure 3: Comparison of the Hcab and nanobody. |
Nanobodies are the variable heavy chain fragment of heavy chain camelid antibodies (Hcab) (Arbabi-Ghahroudi et al., 2017). Although they’re based on the immune system of llamas, synthetic nanobodies, or synbodies, can be produced from plasmids, saving time and money. Nanobodies and synbodies are just as specific for their target antigen as canonical antibodies. They also have the advantage of being significantly smaller, meaning they can recognize epitopes that full antibodies would not physically be able to reach. While a full antibody is ~150 kDa, a nanobody is 12-15 kDa.
Research applications for antibodies
You can use antibodies for experiments in a variety of fields including neuroscience and immunology. They can be used for qualitative and quantitative measurement of protein expression in cell lysates, whole cells, or tissue samples. Many experiments that use antibodies use a primary antibody, which recognizes your protein of interest, and a secondary antibody, which recognizes the primary antibody and provides the method of detection. You’ll also want to use the appropriate controls for your experiment to account for potential non-specific binding of the antibody. Let’s dive in and take a look at some of these applications.
Western blot
Western blots are used to detect the presence of proteins from samples containing a mixture of proteins. The proteins are separated based on molecular weight by SDS-PAGE, and are subsequently transferred to a membrane. A primary antibody recognizing the protein of interest is added, and will bind to that protein on the membrane. Then, addition of a secondary antibody allows the protein to be detected by chemiluminescence or fluorescence. Western blots are frequently used to compare relative levels of protein expression between cell types or treatment conditions. They can be used quantitatively if you also load the gel with appropriate controls for normalizing the protein concentration. To get even more details, check out our article The Basics of Western Blotting.
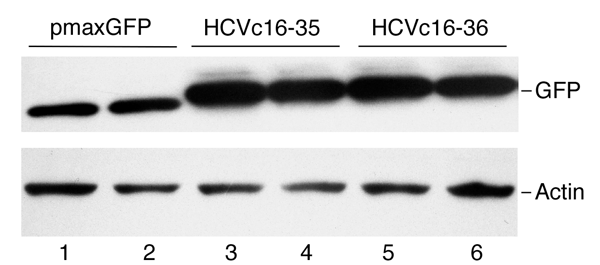 |
| Figure 4: Western blot showing GFP expression in cells that have been transfected with a plasmid encoding GFP. Actin, a common internal control for Western blots, shows that a similar concentration of cell lysate was added to each well. Image from Sun et al., 2010. |
Enzyme-linked immunosorbent assay (ELISA)
Similarly, ELISA is used to detect the presence of a single protein from a heterogeneous mixture - for example, cell lysate or media from cultured cells. ELISA can be used to quantitatively measure the amount of protein present. The reaction takes place in a 96-well plate, where primary and secondary antibodies are used to capture the protein of interest and detect its presence. Then, the amount of light that can pass through each well - the optical density (OD) - of the known and experimental wells is measured.
To find the exact amount of protein in your sample, you’ll compare your samples to a standard curve. This curve is generated using a serial dilution of a known amount of protein that is also detected in the same way. The standard curve that is created from the OD and concentration of the known samples is used to calculate the concentration of protein in the experimental samples. Get more details on ELISAs in our article Antibodies 101: ELISA (Enzyme-linked Immunosorbent Assay).
Immunoprecipitation
Sometimes, instead of just detecting the presence of protein within a sample, you’ll want to extract your protein of interest from the mixture. Antibodies can do this, too! In this method, antibodies bound to microscopic beads grab the protein of interest and pull it out of the solution. Once the unbound protein is washed away, the protein of interest is dissociated from the bead and the antibody using a low-pH buffer. Then, the purified protein can be used in experiments to compare the relative amount of protein present in a series of samples, such as Western blots or ELISAs, or for other experiments to study protein-protein interactions.
Flow cytometry
Flow cytometry is used to detect proteins on the surface of or inside whole cells. Antibodies - typically, primary antibodies that are conjugated to a fluorophore, to avoid the use of secondary antibodies in this experiment - are incubated with a mixture of cells. The technique of using fluorescently-labeled antibodies to detect proteins is referred to as staining. This panel of antibodies is used to both identify the cell subsets of interest from among the mixture and to measure the relative amount of your protein of interest on or in these cells: antibodies can bind to proteins on the cell surface, or the cell membrane can be permeabilized to allow antibodies to bind to intracellular proteins.
The fluorescence signal is detected by the flow cytometer and functions as a proxy for protein expression. Because the signal comes from the antibody-fluorophore conjugate, and is not expressed by the cell itself, there is a lot of flexibility in which proteins you can investigate in each panel. An advantage of flow cytometry is the ability to use multiple fluorescent signals (up to a dozen or more!) to measure the relative or absolute amount of the expression of multiple proteins on the same cell simultaneously as well as compare between samples. Learn more about flow cytometry.
Cell and tissue labeling
Antibodies can be used to visualize tissues (immunohistochemistry) or cells (immunocytochemistry). This could be cells or tissue slices mounted to a slide, or whole organs. Because the tissue architecture is preserved, this technique allows you to not only visualize the presence of specific proteins, but their localization within the cell or tissue as well. Similar to flow cytometry, you’ll use antibodies to stain for markers of one or more specific cell types, as well as for your protein of interest, in order to best contextualize its localization. These could be visualized using fluorophore-conjugated antibodies (i.e., immunofluorescence), or by a colorimetric reaction facilitated by an enzymatic reaction.
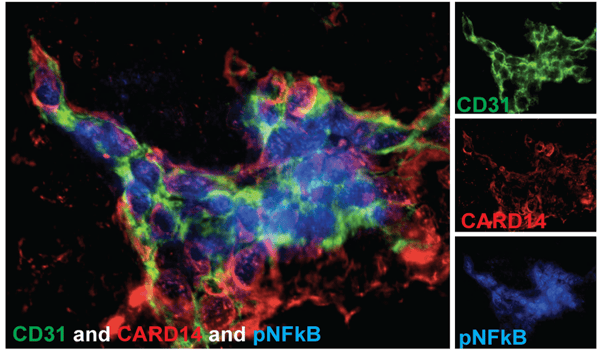 |
| Figure 5: Immunofluorescence staining for CD31 (green), CARD14 (red), and pNFkB (blue) in lesional skin from a psoriasis patient, showing an inflammatory phenotype. Each protein was detected using an unconjugated primary antibody, then visualized using a fluorophore-conjugated secondary antibody or Fab. Image from Harden et al., 2014. |
In vivo uses
For both research and therapeutic purposes, antibodies can be used to affect cellular processes in vivo, and these are referred to as functional grade antibodies. Some of the research and therapeutic uses overlap: antibodies can bind to and inhibit the function of a protein to stop its effect within the body, or they can be used to activate cellular pathways by binding to cell surface receptors. These applications are typically used for investigating signaling pathways or developing therapeutics.
Experimentally, antibodies conjugated to fluorescent tags or other markers can be injected into an animal, to directly label circulating cells or other cells that are accessible from the bloodstream. Then, via a blood draw or organ harvest, the labeled cells can be analyzed using techniques such as flow cytometry.
Conclusion
Whew! Now you know that antibodies can be used for a lot of purposes! I hope you appreciate how many types of antibodies are out there, and how many different research applications they can be used for. Addgene’s Antibody Plasmid Collection includes plasmids that express antibodies, nanobodies, and ScFvs, as well as tools for developing and producing plasmid-based antibodies - and we’re always growing our collection!
References:
Arbabi-Ghahroudi M (2017) Camelid Single-Domain Antibodies: Historical Perspective and Future Outlook. Front Immunol 8: . https://doi.org/10.3389/fimmu.2017.01589
Ayyar BV, Arora S, Ravi SS (2017) Optimizing antibody expression: The nuts and bolts. Methods 116:51–62 . https://doi.org/10.1016/j.ymeth.2017.01.009
Crivianu-Gaita V, Thompson M (2016) Aptamers, antibody scFv, and antibody Fab’ fragments: An overview and comparison of three of the most versatile biosensor biorecognition elements. Biosensors and Bioelectronics 85:32–45 . https://doi.org/10.1016/j.bios.2016.04.091
Cruse JM, Lewis RE, Wang H, Geziena MT, Steven GE, Lorna J. (2004) ANTIGENS, IMMUNOGENS, VACCINES, AND IMMUNIZATION. In: Immunology Guidebook. Elsevier, pp 17–45. https://doi.org/10.1016/B978-012198382-6/50026-1
Wang R, Xiang S, Feng Y, Srinivas S, Zhang Y, Lin M, Wang S (2013) Engineering production of functional scFv antibody in E. coli by co-expressing the molecule chaperone Skp. Front Cell Infect Microbiol 3: . https://doi.org/10.3389/fcimb.2013.00072
Additional resources on the Addgene blog:
- Read about another reagent sharing platform for antibodies, the Developmental Studies Hybridoma Bank
- Learn more about nanobodies
- Watch our Antibodies 101 animation, What is an Antibody?
Resources on Addgene.org:
- Browse Addgene’s Antibody Plasmid Collection
- Find antibodies for neuroscience research from the Trimmer lab
Topics: Antibodies








Leave a Comment