This post was contributed by guest blogger Daniel M. Sapozhnikov, a graduate student in the laboratory of Dr. Moshe Szyf at McGill University.
Some enzymatic fusions are godsends. Tethering dCas9 to the histone acetyltransferase p300 or the DNA methyltransferase DNMT3A allows researchers to easily add biochemical marks to genetic regions from which where they were absent before. But other enzymatic fusions, such as dCas9-TET, can, like the iron ball-and-chain that they resemble, actually impede your investigation into the regulation of your favorite gene. But have no fear - I’m here to introduce you to a simpler, yet more effective, CRISPR-based epigenetic approach (spoiler alert: it’s dCas9… with no modifications). Ready to get started?
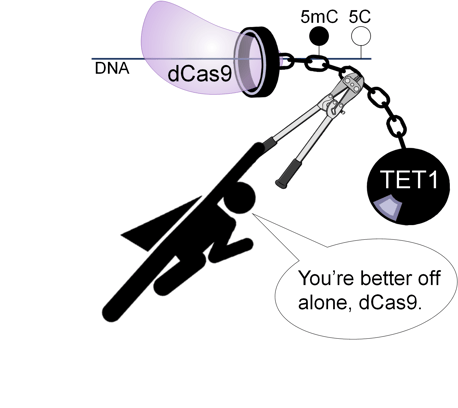 |
|
Figure 1: Break free from the ball and chain! |
Epigenetics nation stuck with methylation correlation
In the 1980s, a lot of things were cool: Walkmans, Back to the Future, and big hair. To what was perhaps a slightly smaller population, something else was also suddenly cool: DNA methylation. In 1983, Andrew Feinberg and Bert Vogelstein showed, for the first time, there were unusual patterns of DNA methylation detected in a human disease: cancer (Feinberg & Vogelstein 1983). The floodgates, as they say, had opened.
In 2021, with over 70,000 articles in PubMed, the idea that DNA methylation is linked to disease has now become much like the 1980s song “Don’t Stop Believin’”; whenever you hear it (whether at a scientific conference or a karaoke bar), you probably want to sigh. The problem is that many of these studies report correlations without clearly testing whether a certain methylation state causes a condition. To understand mechanisms and, eventually, develop treatments, we need tools to specifically edit methylation sites.
Step aside, transcriptionally active dioxygenase
When I started to work on building a CRISPR/dCas9-based tool for DNA demethylation in 2015, none existed. Recently, however, a particular tool has gained widespread popularity – and notoriety – as a tool of choice in the field: dCas9 fused to the enzymatic domain of TET1 or TET2 (Morita et al. 2016, Xu et al. 2016). But there are a number of drawbacks to this approach:
- TET proteins are not demethylases; they are dioxygenases that oxidize 5-methylcytosine to create rare DNA bases that are recognized by distinct proteins and result in unclear transcriptional consequences (Sprujit 2013).
- Enzymatic domains tethered to dCas9 often interact with DNA independently of dCas9 targeting (Galonska 2018). This means that off-target loci – in neighboring chromatin or throughout the genome – could be affected, leading to unpredicted gene expression changes (Sapozhnikov and Szyf 2021, Charlton et al. 2020).
- Most importantly, TET proteins affect transcription independent of their enzymatic activity. They interact with transcriptional machinery to modify local gene expression, confounding any potential conclusions about the role of methylation (Hrit et al. 2018, Kaas et al. 2013).
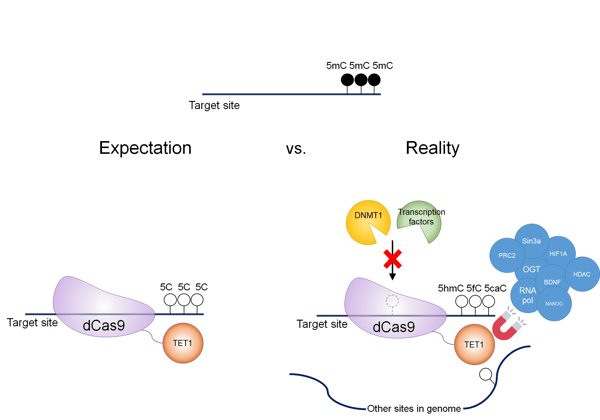 |
|
Figure 2: CRISPR TET-based epigenetic editing. In reality, editing your target site is not as straightforward as expected. TET1 can have many effects independent of dCas9 targeting, including modifying other sites in the genome and directly interacting with transcription machinery. |
Well, I’ve got some good news: complete DNA demethylation can be achieved by dCas9 alone, without TET (or any demethylase) fusion. This may seem counterintuitive: how can a DNA-binding protein with no enzymatic activity cause demethylation?
Dividing cells depend on DNA methyltransferase 1 (DNMT1) to copy methylation marks from parental to daughter strands of DNA. When dCas9 is bound to a target CpG, it physically blocks DNMT1 from that site. So, when DNMT1 methylates the genome during DNA replication, it doesn’t methylate sites blocked by dCas9. Within a few rounds of cell division, the original methylated strand is diluted to undetectable amounts: ergo, DNA demethylation! With optimization, this method can produce nearly 100% demethylation without the confounds inherent to dCas9-TET, allowing one to study the causal relationship between DNA demethylation and gene expression (Sapozhnikov and Szyf 2021).
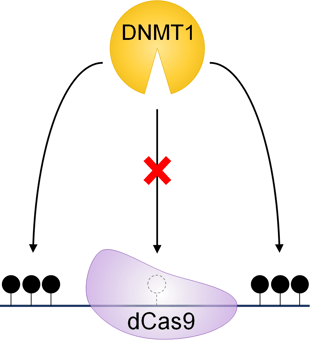 |
|
Figure 3: A simpler approach - dCas9 blocks DNMT1 methylation, leaving the target CpG unmethylated. Adapted from Sapozhnikov and Szyf 2021. |
Let me leave you with a few practical considerations:
- Design your gRNA to overlap the CpG of interest. Having the CpG directly inside the gRNA sequence is a great guarantee that DNMT1 will be blocked (within PAM works, too), but if this is not possible due to gRNA design restrictions, anything within 5 base pairs should work well, and up to 10 base pairs away (especially 3’ of PAM) might work too.
- High dCas9 and gRNA expression are key for complete DNA demethylation – tight binding is crucial for dCas9 to interfere with DNMT1 even through replication.
- The dCas9 also needs to be removeable, otherwise it will continue to block that unmethylated CpG so that nothing else can interact with it. Remember, the goal of this experiment is to expose an unmethylated CpG to behave naturally in the nuclear environment. One good system to balance these needs is the lentiviral-based expression of floxed dCas9 so that it may later be excised by Cre recombinase. Addgene has several great options: pLV hUbC-dCas9-T2A-GFP for removable dCas9 expression and pLM-CMV-R-Cre. Selection for cells stably and highly expressing these constructs can be achieved by fluorescence-activated cell sorting, and stable gRNA expression can be achieved using lentiviral backbones expressing blasticidin or puromycin resistance genes.
It’s that simple! Good luck and happy editing!
 Thank you to our guest blogger!
Thank you to our guest blogger!
Daniel M. Sapozhnikov is a graduate student in the laboratory of Dr. Moshe Szyf at McGill University, working to develop CRISPR-based tools for epigenetic editing.
References and resources:
Additional Resources on the Addgene blog
Resources at Addgene.org
- Read our CRISPR Guide to Epigenetics
References
Charlton J, Jung EJ, Mattei AL et al. (2020) TETs compete with DNMT3 activity in pluripotent cells at thousands of methylated somatic enhancers. Nat Genet 52, 819–827. https://doi.org/10.1038/s41588-020-0639-9.
Feinberg A, Vogelstein B (1983) Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 301, 89–92. https://doi.org/10.1038/301089a0.
Galonska C, Charlton J, Mattei AL et al (2018). Genome-wide tracking of dCas9-methyltransferase footprints. Nat Commun 9, 597. https://doi.org/10.1038/s41467-017-02708-5
Hrit J, Goodrich L, Li C, et al. (2018) OGT binds a conserved C-terminal domain of TET1 to regulate TET1 activity and function in development. eLife 7:e34870. https://doi.org/10.7554/eLife.34870.
Kaas GA, Zhong C, Eason DE, Ross DL, Vachhani RV, Ming G-L, King JR, Song H, Sweatt JD (2013) TET1 Controls CNS 5-Methylcytosine Hydroxylation, Active DNA Demethylation, Gene Transcription, and Memory Formation. Neuron 79, 6 P1086-1093. https://doi.org/10.1016/j.neuron.2013.08.032.
Morita S, Noguchi H, Horii T et al. (2016) Targeted DNA demethylation in vivo using dCas9–peptide repeat and scFv–TET1 catalytic domain fusions. Nat Biotechnol 34, 1060–1065. https://doi.org/10.1038/nbt.3658.
Sapozhnikov DM, Szyf M (2021) Unraveling the functional role of DNA demethylation at specific promoters by targeted steric blockage of DNA methyltransferase with CRISPR/dCas9. Nat Commun 12, 5711. https://doi.org/10.1038/s41467-021-25991-9.
Spruijt CG, Gnerlich F, Smits AH et al. (2013) Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell 152, 5 P1146-1159. https://doi.org/10.1016/j.cell.2013.02.004.
Xu X, Tao Y, Gao X et al. (2016) A CRISPR-based approach for targeted DNA demethylation. Cell Discov 2, 16009. https://doi.org/10.1038/celldisc.2016.9.
Topics: CRISPR, Cas Proteins






Leave a Comment