Since its early years, the light microscope has been a powerful tool in almost all disciplines. In biology, innovative new imaging approaches are constantly being developed which combine computational processing with biological protocols and revolutionary physical approaches. However, with each new technical achievement, the complexity of the device increases, as does the price. With this increased complexity comes the need (and/or opportunities) to modify the device for individual experiments.
Manufacturers rarely provide the construction plans or CAD (computer aided design) drawings of their devices, which makes it very difficult to adapt the device to one's own samples and limits the use of microscopy in open science. But the new field of open-source microscopy seeks to change this. This recent push gives users the chance to understand the "black box" provided by the manufacturers and enables customized experiments. Projects such as the OpenFlexure Microscope (Collins, et. al., 2020), the miCube, SQUID (Hongquan, et. al, 2020) or openUC2 (Diederich et. al., 2020) aim to make microscopy as a tool accessible to practically everyone.
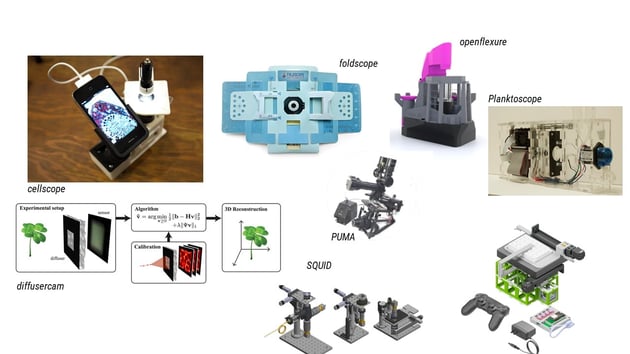
|
| Fig. 1: Open source hardware builds |
Since I started my PhD, I have been convinced that most microscopy techniques can be replicated with commercially available consumer-grade components. In this field, often referred to as 'frugal science', existing components are adapted and combined with creativity and feedback from the community to create state-of-the-art devices for research. There are a number of new platforms (such as openUC2, which I am a co-founder of), that aim to support the community in developing better, more sustainable, easily sharable products.
Open-Source Microscopy
Sharing Hardware vs. Sharing Software
In the field of microscopy, where image processing algorithms for analyzing time-lapse images, quantitatively evaluating antibody staining, or counting cells are now routine, open-source software projects that actively engage with the community are particularly successful. Machine-learning denoising of raw data (CARE), segmentation of imaging data (ILASTIK), or the popular plugin-based image process framework FIJI are just three of the constantly growing number of "community-driven" software that make everyday microscopy much easier. On platforms such as Github, users can download the software for use, or they can modify and optimize the code, find and fix security-critical bugs, and contribute to constant updates of the software.
In hardware, this process is often much more involved. Challenges such as the provision of an accessible supply chain for parts that are listed in the bill of material (BOM) of an assembly, the feasibility of manufacturing, and subsequent assembly can be difficult to overcome. Universities may not have the opportunity to buy parts through low-cost sources such as Amazon, Ebay, or Aliexpress, which makes it difficult to develop accessible open-source hardware.
Projects such as the OpenFlexure microscope or open-source modular optical toolbox openUC2 are trying to overcome this hurdle by taking these limitations into account in the development process. Widely used, commercially available tools such as the Raspberry Pi, the Arduino, or 3D printers are key to enabling this work. Sharing open-source plans that rely on such tools has allowed the community to regularly replicate designs worldwide. Distributing hardware via globally established makerspace networks and training users via workshops, seminars, or hackathons greatly increases accessibility. Through this approach, the Planktoscope, a portable device for studying plankton using flow cells, has become an established alternative to high-priced commercial devices.
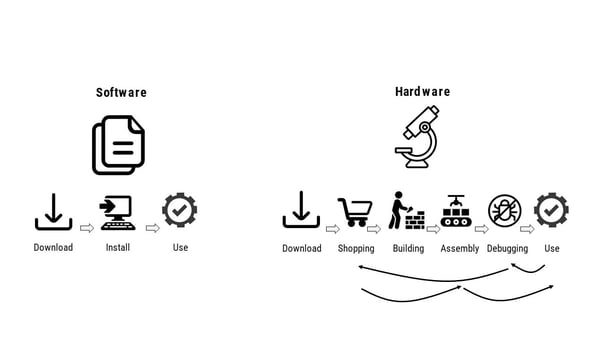
|
| Fig. 2: Workflow for open software (left) vs. open hardware (right). |
Taking this all together, the essential ingredients of open-source microscopy are as follows:
Detailed documentation, which facilitates the reproduction of the structure in a similar way to the biological protocol, and assembly designs based around the use of commercially available tools and components.
Rapid prototyping techniques such as 3D printing, on-demand CNC milling, and PCB manufacturing. These newly developed techniques are the key enablers for making assemblies open-source. The sharing of digital representations of the 3D models, which in many cases can be produced quickly with the printer, act here as a kind of "teleporter," since a part developed in Germany, for example, can also be printed quickly in Madagascar.
Platforms that allow for on-going creativity with the construction plans. Appropriate open-source licenses for hardware (e.g. CERN OHL), software (e.g. GPL v3, MIT), and documentation (e.g. CC-BY) allow users outside the project to modify the raw and design files, allowing the device to take on a community-driven life of its own.
Nevertheless, there are still many open questions to be clarified, especially with regard to licensing models. Direct sharing of work from universities and research centers provides a potential conflict between traditional IP-based models and the use of taxpayer money to support this research. But there is a move towards more open research. The EU, for example, is calling for the opening of sources within the next few years, with the goal of creating more transparent and reproducible science in the long run (Hohlbein, et. al., 2022). A more open way of communicating in science brings stronger networking across laboratory and disciplinary boundaries and can allow for innovative organizations and companies to provide problem-solving platforms.
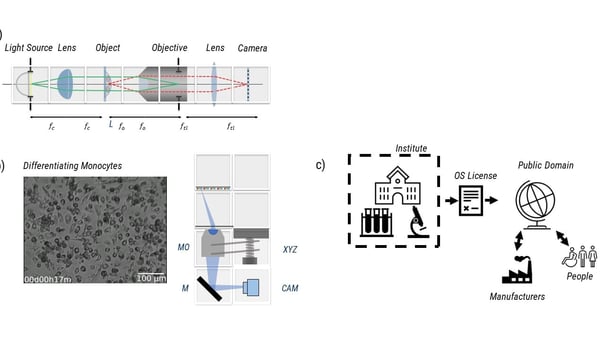
|
| Fig. 3: Different workflows involved in sharing open hardware. Upper left panel shows the path of light through a microscope. Lower left panel shows an image of differentiating monocyte with planes and hardware to it. Right panel shows workflow of open hardware. |
Conclusion
Addgene is a great example of how sharing of data and molecular tools can lead to faster and more efficient science. Researchers around the world don't have to start from scratch every time to pursue their scientific curiosity but can literally put their work on the shoulders of giants. The still relatively young movement of sharing open hardware has a similar goal. The sharing of modules, ideas, and blueprints has the potential not only to accelerate scientific progress, but also to enable science in parts of the world that are otherwise cut off from the process.
About the author: Benedict Diederich received his PhD from the Heintzmann Lab at the Leibniz IPHT Jena. He is co-founder of the open-hardware startup “openUC2” and focuses on bringing cutting-edge research to everybody by relying on tailored image processing and low-cost optical setups.
References
Joel T. Collins, Joe Knapper, Julian Stirling, et. al, "Robotic microscopy for everyone: the OpenFlexure microscope," Biomed. Opt. Express 11, 2447-2460 (2020)
Hongquan Li, Deepak Krishnamurthy, Ethan Li, Pranav Vyas, Nibha Akireddy, Chew Chai, Manu Prakash, "Squid: Simplifying Quantitative Imaging Platform Development and Deployment." bioRxiv. 2020.
Diederich, B., Lachmann, R., Carlstedt, S. et al. A versatile and customizable low-cost 3D-printed open standard for microscopic imaging. Nat Commun 11, 5979 (2020). https://doi.org/10.1038/s41467-020-19447-9
Hohlbein, J., Diederich, B., Marsikova, B. et al. Open microscopy in the life sciences: quo vadis?. Nat Methods 19, 1020–1025 (2022). https://doi.org/10.1038/s41592-022-01602-3
Topics: Scientific Sharing, Imaging, Open Science





Leave a Comment