Picture this: you’ve been assigned an exciting new project aimed at understanding how a critical cellular pathway is regulated. You’ve read all the background papers you could get your hands on, formulated a hypothesis, and planned out your key experiments. Unsurprisingly, many of these experiments require antibodies. You run a quick internet search for the required antibodies and find….hundreds of options. With limited time and budget you cannot test them all, so how do you know which ones to try?
Read on and we will provide you with some advice on the antibody selection process.
Antibodies are not one size fits all
When choosing an appropriate antibody first look at the application-specific data provided on the manufacturer’s website. Because of how they are developed, antibodies rarely work for every assay. Antibodies that are generated against a linear antigen tend to work very well for applications that denature a protein to its primary (linear) structure using heat or acid, for example in a Western blot. If you were to try and use such an antibody in an assay for a native (folded) protein, such as immunoprecipitation, the epitope may not be accessible for binding and the antibody will not work. Whenever possible, choose an antibody that has been validated in your specific application.
Consider also that there may be variations within an application. Take immunohistochemistry (IHC), for instance. While an antibody may be validated for IHC, not all IHC experiments are the same. Differences in sample processing, fixation, antigen-retrieval, and permeabilization will influence how the antibody interacts with its target and consequently the quality of the stain. Choose the antibody that was successfully used in conditions that are the most similar to yours. Take a look at the publications cited on the manufacturer’s website, which may contain more detailed data on the assay methods. You might find that while the website highlights a variation of your application, a publication successfully used it in a similar way as you plan to.
To secondary or not to secondary?
The application will also determine whether you should use an antibody that is directly conjugated to a detection substrate, such as a fluorophore, or a conjugated secondary antibody that will bind to your primary antibody. If you are trying to visualize the precise location of a protein in a tissue section then a directly-conjugated antibody will provide better resolution as these immune complexes are smaller than a primary:secondaries:conjugates complex. If you are trying to visualize a protein in low abundance on a Western blot, a secondary antibody is ideal as it will help to amplify the signal. Some applications use either direct (primary-conjugated) or indirect (secondary) methods. In those cases, remember that a secondary should be used when signal amplification is needed.
Species
Next, check the species that the antibody cross-reacts with. Ideally you will find an antibody that has been validated in the species your protein of interest comes from. If one is not available, don’t lose hope! It could still work. Take a close look at the antigen or antigenic fragment that was used to develop the antibody. Is this protein or protein region highly conserved across species? If the proteins are highly homologous between your target species and the species antibody cross-reacts to, then there is a good chance that the antibody could meet your needs.
Multiplex experiments
Finally, consider whether you will be using other antibodies in the experiment. If you are planning to use multiple antibodies in a single experiment then you will need to make sure that you can easily distinguish each different type of antibody and therefore each target protein. This can be accomplished by using a panel of primary antibodies conjugated to different fluorophores or a panel of primaries that was raised in different species or are unique isotypes. If every antibody in the assay is a unique isotype or species, then you can choose different fluorophore-conjugated secondary antibodies specific to each isotype or species to easily discern the proteins in your assay.
Narrowing the field
After working your way through the above steps, you may find that you still have a number of options. If this is the case, you can narrow them down in a variety of ways. First, review the citations. An antibody that has been cited thousands of times may be a safer bet than one that has never been used in a publication. Talk with your peers. If someone in your field has used the antibody before you will not only have peace of mind as you make your purchase but also someone to turn to for advice if you need to troubleshoot. What do you know about the vendor and the vendor’s technical support? An antibody from a vendor that you use frequently, with a responsive and knowledgeable technical support team, is a safer bet than one from a large distributor that may not have direct experience with the product. Note that many antibodies are sold under different names through multiple vendors, so look at epitope target, source, and other information to make sure you’re not trying to choose between the same antibody sold under different names.
If all else is equal, feel free to make the decision by price point, shipping time, or other factors. Just remember that these are secondary considerations to validation, application specificity, and experimental considerations - no other factors can make up for an antibody that doesn't work for your application.
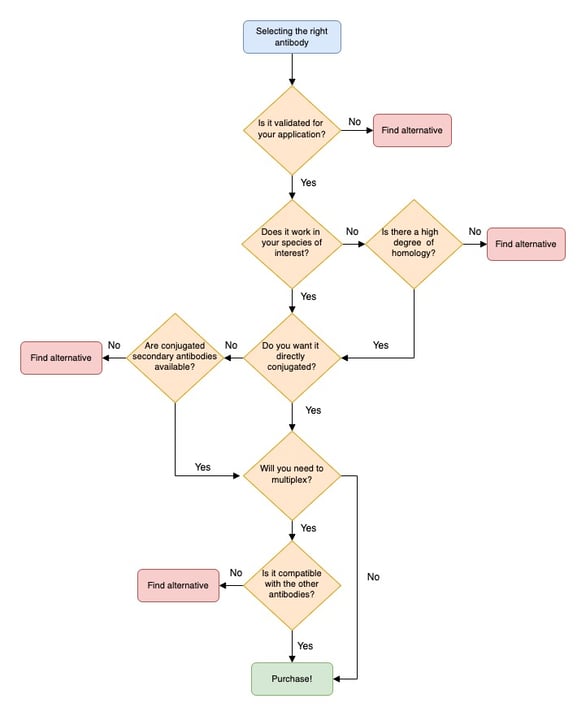
|
| Fig. 1: Antibody Selection Flow Chart |
What does validated mean anyway?
You now know that application validation is the most important factor in antibody selection, but what does that mean? Not all validation is equal and you should think critically about what data is and is not shown on an antibody website. In 2016, the International Working Group for Antibody Validation proposed 5 pillars for antibody validation.
The 5 pillars include genetic strategies that test expression in knock-out or knock-down down tissues and cell lines; orthogonal strategies that compare antibody assay results with those of antibody-independent assays such as mRNA data from transcriptomics; independent antibody strategies that compare the staining patterns of multiple antibodies against the same target; tagged protein strategies that express a tagged version of the antigen and compare staining of the antibody to that of the anti-tag antibody; and immunoprecipitation-mass spectrometry strategies that isolate an antibody’s immune complexes and confirm interacting partners by mass spectrometry (Uhlen, 2016). Be sure that the antibody you choose has been validated using one or more of the proposed pillars.
Remember, validation does not mean that the antibody is guaranteed to work. Antibody-based assays are complex and antibodies may perform differently in different samples and experimental conditions. Be prepared to troubleshoot and be sure to include proper positive and negative controls as you get your assays up and running.
Ed.'s note: More questions on antibody validation? Check out Antibodies 101: Validation!
Download our antibody selection flow chart.
References and Resources
Additional Resources on the Addgene blog
The Basics of Western blotting
Antibodies 101: Introduction to Antibodies
Antibodies 101: Epitope availability
References
Uhlen M, Bandrowski A, Carr S, Edwards A, Ellenberg J, Lundberg E, Rimm DL, Rodriguez H, Hiltke T, Snyder M, Yamamoto T (2016) A proposal for validation of antibodies. Nat Methods 13:823–827
Topics: Antibodies






Leave a Comment