This post was contributed by guest blogger, Robert Orr, who recently received a Ph.D. in Biology and Biotechnology from Worcester Polytechnic Institute.
What is RNAi?
The loss-of-function (LOF) experiment functions as the building block of our understanding of complex biological processes. Many tools exist to perturb biological function in a direct or unbiased way at the DNA, RNA, or protein level. The “correct” choice of tool requires careful balancing of the inherent advantages and limitations of any technique in the context of the biological question. For example, while gene knockouts have long been considered the “gold-standard” for LOF studies, the high gene copy number found in plants makes traditional knockouts unattractive from a practical perspective. Therefore, techniques that function downstream of DNA, such as RNA interference, can reversibly exert their effect independent of gene copy number.
RNA interference (RNAi) is a conserved eukaryotic process where approximately 20-30 nucleotides of double-stranded RNA (dsRNA) results in downregulation, or “silencing,” of any gene that contains sequence complementary to the dsRNA (Wilson and Doudna, 2013).
RNAi consists of two non-exclusive gene silencing pathways: transcriptional and post-transcriptional gene silencing (PTGS). In either pathway, short double-stranded RNA fragments (siRNAs) are produced from long dsRNA via an RNase III enzyme named Dicer (Figure 1). The plant kingdom has expanded its repertoire of Dicer enzymes and these different Dicer-like enzymes are involved in the biogenesis of different small RNAs (Figure 1). These siRNAs can exert their gene silencing effect through targeted mRNA degradation, blocking translation, or chromatin modifications of complementary gene sequences (Figure 1). As a result, gene silencing can be easily induced by exogenous application of long or short dsRNA. However, it is important to note that naked and unmodified dsRNA elicits immunogenic reactions that complicate the effectiveness and interpretation of RNAi. Fortunately, researchers studying plants and other organisms lacking this innate immunity need not worry and have benefited from the ease of use in dsRNA applications.
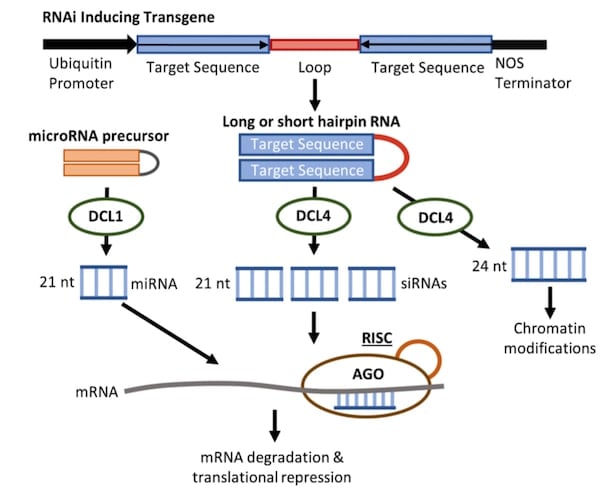 |
| Figure 1: Schematic of an RNAi-inducing transgene. Typically, the desired target of RNAi is expressed as inverted repeats separated by a loop region that facilitates the formation of a hairpin RNA structure, resulting in processing by the endogenous RNAi machinery. Alternatively, endogenous miRNA precursors can be engineered to created artificial miRNAs. The processed miRNA or siRNAs then silence the target gene(s) either transcriptionally or post-transcriptionally. In plants, different dicer-like enzymes (DCL) process the dsRNA into specific types of interfering RNAs. |
RNAi vs. CRISPR: Complementary techniques
It is fair to say that CRISPR has revolutionized molecular biology and has enabled functional studies in unprecedented ways. With the meteoric rise and diversity of CRISPR technologies, one might incorrectly relegate RNAi to history. The straightforwardness of RNAi is unmatched—unlike CRISPR experiments that require exogenous protein, successful induction of RNAi only requires the presence of dsRNA. In plants, CRISPR technologies are predominantly used to disrupt function at the level of DNA, thereby limiting CRISPR’s effectiveness at high copy numbers and genomic loci with low accessibility. Furthermore, gene knockout strategies cannot be used to investigate essential genes.
Historically, gene silencing methods were thought to be afflicted by pervasive off-target effects. This conclusion was based upon numerous phenotype discrepancies between gene silencing and knockout methods of the same gene. However, recent work has called this prevalent assumption into question. Specifically, CRISPR-generated mutants believed to be knockouts can maintain partial function due to the use of exon skipping, alternative start codons, and/or the phenomenon of transcriptional adaptation (Smits et al., 2019; Sztal and Stainier, 2020). Additionally, a study that directly compared CRISPR to RNAi in the context of a high throughput survival screen found that either technology was equivalently precise, but each method recovered different genes known to be required for viability (Morgens et al., 2016). Only upon combining the data generated from both methods did a more comprehensive gene list emerge that accurately reflected the scope of genes known to be essential for cell growth. Together, this excellent work highlights the importance of using orthogonal methods to confidently conclude the function of a gene of interest.
Tools for RNAi
Arguably the greatest advantage of RNAi is the straightforward nature of the technique—all that is required dsRNA complementary to your gene or genes of interest to trigger RNAi activation.
A typical RNAi experiment can be divided into three fundamental steps:
- creation of the RNAi trigger
- delivering the RNAi trigger
- characterizing the RNAi phenotype.
Although all these steps are areas of active development, the first step necessarily dictates the direction and scope of the subsequent two. Consequently, I will focus on different approaches and considerations present at the first stage of an RNAi-based investigation.
Creation of the RNAi trigger requires a few initial considerations, such as the starting material (DNA-based vector or purified RNA) and the initial state of the dsRNA upon activating the RNAi machinery (long dsRNA, typically hairpin RNA, or artificial microRNA). Using a DNA template for the RNAi trigger is the most prevalent method due to ease of use and capacity to generate a long-term RNAi phenotype.
Long hairpin RNA
The easiest approach to induce RNAi involves the expression of long hairpin RNA (hpRNA). This method is facilitated by DNA constructs that enable insertion of ~400 bp complementary to your gene target as inverted repeats (Figure 1). With this technique, multiple genes can be simultaneously silenced if a consensus sequence is used (~90% identity between targets) or multiple target sequences are concatenated. hpRNA has been extensively used in plant research and many different constructs are available to the community. Each construct has unique features, such as inducible expression of the hpRNA and/or fusing your gene target sequence to a fluorescent reporter sequence (discussed below). Examples of these constructs include pGAPi, which allows easy creation of long hpRNA with Gateway-based cloning and straightforward positive selection of plants undergoing RNAi, and LIIbeta F 1-2 RNAi, which allows assembly of intron-spliced hpRNAs through Golden Gate cloning.
Artificial micro RNA and short hairpin RNA
The greatest limitations of the long hpRNA approach are thought to be off-target effects and variable silencing efficiency. To address these shortcomings, both short hpRNAs (shRNAs) and artificial micro RNAs (amiRNAs) are frequently used. Although similar, amiRNAs are engineered from endogenous precursors of microRNAs found in the genome, whereas shRNAs are exogenously derived. Importantly, instead of the assortment of siRNAs generated from a long hpRNA trigger, only a single ~21-nt interfering RNA is produced using shRNA/amiRNA. Although sh- and amiRNAs are likely more specific than long hpRNAs, they must be individually engineered and empirically evaluated for their gene silencing efficacy, thereby requiring a substantial time investment.
Once the target mRNA is cleaved, the cleaved mRNA can induce further rounds of RNAi. This process, known as “transitivity,” calls into question any claims about specificity of a given RNAi technique. While transitivity enhances the potency of RNAi, it can also increase the occurrence of off-target effects. Nevertheless, amiRNA-based RNAi experiments routinely display highly specific and robust gene silencing (following initial optimization of the amiRNA). Although downstream work is required, several vectors can be found at Addgene that could serve as an excellent start (#22988, #137884). Both vectors allow the creation of amiRNAs based upon miRNA precursors of rice and Arabidopsis species, respectively.
Short hairpin RNA (shRNA) is conceptually similar to the microRNA precursor mentioned above and is a popular RNAi trigger in the mammalian research community. Fortunately, many tools for shRNA in mammals are available at Addgene.
Identifying actively silenced plants
Once you’ve introduced your RNAi into your organism, how do you identify the actively silenced ones? Here, I’ll cover some screening methods to identify them.
Visual screening
Regardless of the RNAi-trigger type you decide, any RNAi experiment benefits from an easy method to identify and quantify actively silencing plants. Non-destructive methods, most popularly with a fluorescent reporter, directly couple silencing of your gene target to silencing of the reporter. This is achieved at the level of the DNA construct, where your RNAi-target is fused to the DNA backbone that contains a target sequence against the fluorescent reporter in an inverted repeat orientation. Therefore, a single hairpin dsRNA is produced that contains sequence against your gene(s) of interest and the reporter. This approach greatly enhances experimental throughput, as silencing plants can be easily identified and have readily observable phenotypes quantified. Fluorescent reporter-based RNAi constructs have been designed and widely implemented in plants, such as in the model flowering plant Arabidopsis thaliana (Li et al., 2013; Zhang et al., 2018) and the model bryophyte Physcomitrium (Physcomitrella) patens (Bezanilla et al., 2005; Nakaoka et al., 2012).
A new positive selection-based RNAi method
Until now, all DNA-based RNAi experiments typically involved antibiotic selection to test for the presence of the RNAi transgene, followed by a fluorescence readout of RNAi activity. As a plant researcher, the traditional hpRNA and visual screening method was limited in two substantial ways: variable silencing efficiency and labor-intensive processes to isolate RNAi plants. With a visual reporter of RNAi, phenotypes, such as morphological parameters, can be easily scored. However, determining the transcript or protein abundance of your specific target required physical isolation of the visually identified subpopulation. For traditional cell culture, enriching your active RNAi subpopulation can be easily achieved through fluorescence-activated cell sorting (FACS). This is not possible in plants outside of limited, single cell protoplast analysis. Instead, microscopic plants must be visually identified as undergoing RNAi through the fluorescent reporter and manually extracted. This process is tedious and time consuming, as RNAi mutants with a reduced growth phenotype will require additional plants to have enough tissue for downstream characterization.
To simultaneously address both limitations of the traditional RNAi approach, myself and others in the Luis Vidali lab designed a new RNAi method. Instead of using a fluorescent output to monitor RNAi, we engineered a positive selection reporter whereby survival itself is indicative of active gene silencing. We achieved this by creating constructs that directly fused any gene targeting sequence to a targeting sequence for the adenine phosphoribosyltransferase (APT) gene, which is conserved in many organisms. When supplemented with adenine analogs such as 2-fluoroadenine (2-FA), organisms with functional APT will convert 2-FA to cytotoxic nucleotides, resulting in death. In the presence of 2-FA, potent gene silencing of APT is required for survival. By coupling silencing of APT and any gene target of interest, only organisms that are actively silencing to a sufficient degree remain alive. This likely reduces variability in the RNAi output and trivializes downstream characterization, as the entire culture can be harvested instead of microscopic isolation. The plasmids required to conduct an APT-based RNAi experiment are now available on Addgene. These include the construct that allows for easy insertion of any target sequence, as well as essential control plasmids.
Many thanks to our guest blogger Robert Orr!
 Robert Orr recently earned his Ph.D. in Biology and Biotechnology from Worcester Polytechnic Institute where he investigated how plants achieve polarized cell growth in the lab of Luis Vidali. For his next adventure, he will join the lab of Darren Gilmour at the University of Zurich. He is fascinated by how organisms exquisitely and robustly self-organize at the cellular and multicellular scale.
Robert Orr recently earned his Ph.D. in Biology and Biotechnology from Worcester Polytechnic Institute where he investigated how plants achieve polarized cell growth in the lab of Luis Vidali. For his next adventure, he will join the lab of Darren Gilmour at the University of Zurich. He is fascinated by how organisms exquisitely and robustly self-organize at the cellular and multicellular scale.
References
Bezanilla M, Perroud P ‐F., Pan A, Klueh P, Quatrano RS (2005) An RNAi System in Physcomitrella patens with an Internal Marker for Silencing Allows for Rapid Identification of Loss of Function Phenotypes. Plant Biology 7:251–257 . https://doi.org/10.1055/s-2005-837597
Li J-F, Chung HS, Niu Y, Bush J, McCormack M, Sheen J (2013) Comprehensive Protein-Based Artificial MicroRNA Screens for Effective Gene Silencing in Plants. Plant Cell 25:1507–1522 . https://doi.org/10.1105/tpc.113.112235
Morgens DW, Deans RM, Li A, Bassik MC (2016) Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat Biotechnol 34:634–636 . https://doi.org/10.1038/nbt.3567
Nakaoka Y, Miki T, Fujioka R, Uehara R, Tomioka A, Obuse C, Kubo M, Hiwatashi Y, Goshima G (2012) An Inducible RNA Interference System in Physcomitrella patens Reveals a Dominant Role of Augmin in Phragmoplast Microtubule Generation. Plant Cell 24:1478–1493 . https://doi.org/10.1105/tpc.112.098509
Smits AH, Ziebell F, Joberty G, Zinn N, Mueller WF, Clauder-Münster S, Eberhard D, Fälth Savitski M, Grandi P, Jakob P, Michon A-M, Sun H, Tessmer K, Bürckstümmer T, Bantscheff M, Steinmetz LM, Drewes G, Huber W (2019) Biological plasticity rescues target activity in CRISPR knock outs. Nat Methods 16:1087–1093 . https://doi.org/10.1038/s41592-019-0614-5
Sztal TE, Stainier DYR (2020) Transcriptional adaptation: a mechanism underlying genetic robustness. Development 147:dev186452 . https://doi.org/10.1242/dev.186452
Wilson RC, Doudna JA (2013) Molecular Mechanisms of RNA Interference. Annu Rev Biophys 42:217–239 . https://doi.org/10.1146/annurev-biophys-083012-130404
Zhang N, Zhang D, Chen SL, Gong B-Q, Guo Y, Xu L, Zhang X-N, Li J-F (2018) Engineering Artificial MicroRNAs for Multiplex Gene Silencing and Simplified Transgenic Screen. Plant Physiol 178:989–1001 . https://doi.org/10.1104/pp.18.00828
Additional resource on the Addgene blog
- Browse our blog posts about plant biology
- Check out our CRISPR blog posts
Resources on Addgene.org
- Find tools for mammalian RNAi
- Browse all plasmids for RNAi
- Check out Addgene's plasmids and resources for plant biology
Topics: Plant Biology, Other Plasmid Tools, Plasmids






Leave a Comment