Working on the scientific support team at Addgene, I get to see everything that can go wrong with a plasmid. One of the most frequent problems that I help troubleshoot involves viral vectors and plasmids with repeating sequences. Repeating sequences within a plasmid can undergo intramolecular DNA recombination. This naturally occurring process is mediated by recombinases like RecA and RecBCD in bacteria. When two repeating elements love each other very much undergo recombination, two smaller plasmids are formed.
Viral vectors are prone to recombination
In lentiviral vectors, recombination often occurs between the long-terminal repeats (LTRs). After recombination, one resulting plasmid carries the transgene of interest (aka the viral genome) and the other contains the bacterial selection marker and origin of replication. During bacterial culture, only the plasmid with the antibiotic resistance gene and origin will replicate, and the transgene will be lost. Over time, bacteria containing the recombined vector backbone will also outgrow bacteria that contain the full length plasmid.
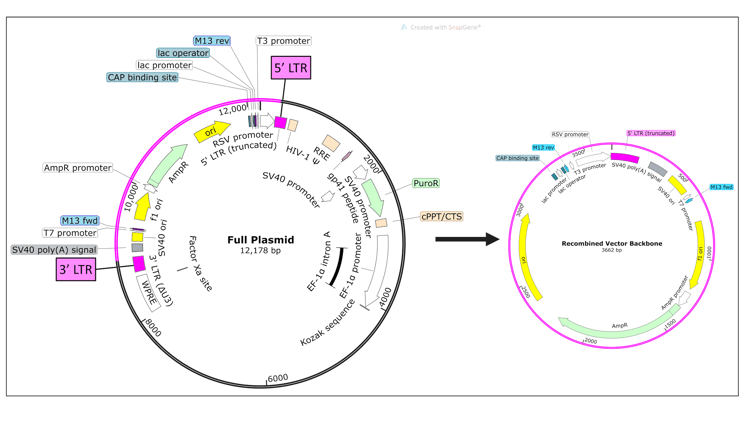 |
| Figure 1: The full lentiviral plasmid is expected to be >12 kb. However if it recombines at the 5’ and 3’ LTR regions, it splits into two smaller plasmids. The ~3.7 kb recombined vector backbone contains the antibiotic resistance gene (AmpR) and origins of replication (ori and f1 ori) which allow it to replicate. |
How can I prevent recombination in my samples?
Plasmid recombination can affect viral vectors (LTRs and ITRs), plasmids with multiple polyA sites, and other plasmids that contain repeating elements. Knowing how to amplify recombination-prone plasmids and testing your prep can save you the headache of troubleshooting failed experiments.
Pick the right strain
While common E. coli strains like DH5-alpha are recA-, some strains have been engineered to further reduce recombination. Recombinase-deficient strains like Stbl2, Stbl3, and NEB stable E. coli reduce the rate of recombination in samples.
For a more detailed comparison of these cloning strains, browse the table of common gene mutations in our Plasmids 101: Common Lab E. coli Strains blog post. Check yourself before you recA yourself.
Optimize Growth Conditions
To reduce recombination, it is important to not overgrow the plasmid at 37°C. Some researchers find that using a 30°C incubation temperature to grow bacteria more slowly helps reduce the risk of recombination. Look for the “Growth in Bacteria” section on a plasmid webpage for any specific recommendations from the depositing lab.
If your vector is large (>10 kb) or has a low copy number, try following protocols for low copy plasmids. For example, using a longer growth period (up to 24 hr), reducing the concentration of antibiotics (ie. 50 μg/mL Amp instead of 100 μg/mL Amp), or using more nutrient rich media can improve growth and DNA yield.
Test multiple, small colonies
After streaking LB agar plates, you may notice that there are both large and small colonies. Bacteria containing the recombined product tend to grow faster, so we recommend picking the smaller colonies which are more likely to carry the full plasmid. Chakiath & Esposito (2018) noticed that in Stbl3 bacterial cultures with lentiviral vectors, the flat white colonies contained the full plasmid, whereas the round tan colonies that look similar to normal E. coli cultures contain the recombined vector backbone.
Test multiple colonies before saving a glycerol stock to ensure the colony you selected contains the full plasmid. A diagnostic digest is one of the easiest and clearest ways to determine if your sample has recombined. Choose a restriction enzyme that will give you different band patterns for the recombined vector backbone vs. the full length plasmid. If you find that your bacterial cultures contain a mix of full length plasmid and the recombined vector backbone, you can try re-streaking for single colonies and testing multiple preps. In some cases, we suggest gel extraction to isolate the full, intact plasmid. Please see our gel extraction protocol for more details.
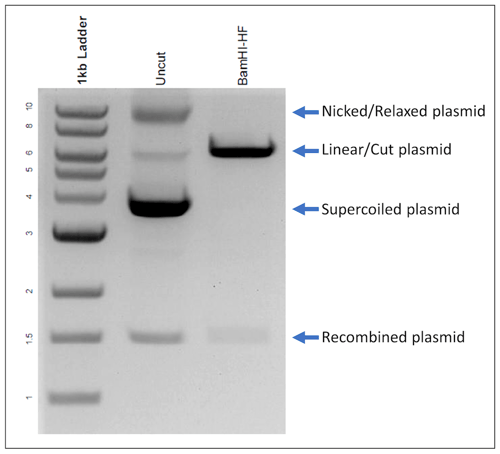
|
|
| Figure 2: An uncut plasmid is expected to form 3 bands: nicked, linear, and supercoiled. Here we find a smaller, recombined plasmid around 1.5 kb. When cut with BamHI, the expected linear plasmid is present, but the recombined plasmid is still visible. This indicates that the DNA prep contains a mixture of the full plasmid and a smaller percentage of the recombined vector backbone. |
Ready, set, grow! Our 5 Tips for Troubleshooting Viral Transductions will help with the next steps.
References and resources
References
Al-Allaf FA, Tolmachov OE, Zambetti LP, Tchetchelnitski V, Mehmet H (2012) Remarkable stability of an instability-prone lentiviral vector plasmid in Escherichia coli Stbl3. 3 Biotech 3:61–70. https://doi.org/10.1007/s13205-012-0070-8
Trepotec Z, Geiger J, Plank C, Aneja MK, Rudolph C (2019) Segmented poly(A) tails significantly reduce recombination of plasmid DNA without affecting mRNA translation efficiency or half-life. RNA 25:507–518. https://doi.org/10.1261/rna.069286.118
Additional resources on the Addgene blog
- Find more molecular biology and viral vector protocols
- Learn about the common lab E. coli strains
- Find tips for viral transduction
Topics: Viral Vector Protocols and Tips





Leave a Comment